MDStudio2 is a program for cartographic analysis of population genetic data and other numerical characters distributed in geographic space. The basis of the analysis consists of methods of interpolation and design of contiguous geographical fields as well as methods of mathematical transformation and statistic analysis of geographic distributions.
2011年12月28日星期三
2011年12月27日星期二
pairwise sequence alignment with R
http://biostar.stackexchange.com/questions/15734/pairwise-sequence-alignment-with-r
Finding Data on the Internet
http://www.inside-r.org/howto/finding-data-internet
What I would like is a nice list of all of credible sources on the Internet for finding data to use with R projects. I know that this is a crazy idea, not well formulated (what are data after all) and loaded with absurd computational and theoretical challenges. (Why can't I just google "data R" and get what I want?) So, what can I do? As many people are also out there doing, I can begin to make lists (in many cases lists of lists) on a platform that is stable enough to survive and grow, and perhaps encourage others to help with the effort.
Here follows a list of data sources that may easily be imported into R.
If an (R) appears after source this means that the data are already in R format or there exist R commands for directly importing the data from R. (See http://www.quantmod.com/examples/intro/ for some code.) Otherwise, i have limited the list to data sources for which there is a reasonably simple process for importing csv files. What follows is a list of data sources organized into categories that are not mutually exclusive but which reflect what's out there.
Economics
UMD:: http://inforumweb.umd.edu/econdata/econdata.html
World bank: http://data.worldbank.org/indicator
Finance
CBOE Futures Exchange: http://cfe.cboe.com/Data/
Google Finance: http://finance.yahoo.com/ (R)
Google Trends: http://www.google.com/trends?q=google&ctab=0&geo=all&date=all&sort=0
St Louis Fed: http://research.stlouisfed.org/fred2/ (R)
NASDAQ: https://data.nasdaq.com/
OANDA: http://www.oanda.com/ (R)
Yahoo Finance: http://finance.yahoo.com/ (R)
Government
Archived national government statistics: http://www.archive-it.org/
Australia: http://www.abs.gov.au/AUSSTATS/abs@.nsf/DetailsPage/3301.02009?OpenDocument
Canada: http://www.data.gc.ca/default.asp?lang=En&n=5BCD274E-1
DataMarket: http://datamarket.com/
Fed Stats: http://www.fedstats.gov/cgi-bin/A2Z.cgi
Guardian world governments: http://www.guardian.co.uk/world-government-data
London, U.K. data: http://data.london.gov.uk/catalogue
New Zealand: http://www.stats.govt.nz/tools_and_services/tools/TableBuilder/tables-by...
NYC data: http://nycplatform.socrata.com/
OECD: http://www.oecd.org/document/0,3746,en_2649_201185_46462759_1_1_1_1,00.html
San Francisco Data sets: http://datasf.org/
U.K. Government Data:http://data.gov.uk/data
United Nations: http://data.un.org/
U.S. Federal Government Agencies: http://www.data.gov/metric
US CDC Public Health datasets: http://www.cdc.gov/nchs/data_access/ftp_data.htm
The World Bank: http://wdronline.worldbank.org/
Machine Learning
Causality Workbench: http://www.causality.inf.ethz.ch/repository.php
Kaggle competition data: http://www.kaggle.com/
KDNuggets competition site: www.kdnuggets.com/datasets/
UCI Machine Learning Repository: http://archive.ics.uci.edu/ml/
Machine Learning Data Set Repository: http://mldata.org/
Microsoft Research: http://research.microsoft.com/apps/dp/dl/downloads.aspx
Million songs: http://blog.echonest.com/post/3639160982/million-song-dataset
Social Networking: http://www.cs.cmu.edu/~jelsas/data/ancestry.com/
Public Domain Collections
Data360: http://www.data360.org/index.aspx
Datamob.org: http://datamob.org/datasets
Factual: http://www.factual.com/topics/browse
Freebase: http://www.freebase.com/
Google: http://www.google.com/publicdata/directory
infochimps: http://www.infochimps.com/
numbray: http://numbrary.com/
Sample R data sets: http://stat.ethz.ch/R-manual/R-patched/library/datasets/html/00Index.html (R)
SourceForge Research Data: http://www.nd.edu/~oss/Data/data.html
UFO Reports: http://www.nuforc.org/webreports.html
Wikileaks 911 pager intercepts: http://911.wikileaks.org/files/index.html
Stats4Stem.org: R data sets: http://www.stats4stem.org/data-sets.html (R)
The Washington Post List: http://www.washingtonpost.com/wp-srv/metro/data/datapost.html
Science
Agricultural Experiments: http://www.inside-r.org/packages/cran/agridat/docs/agridat (R)
Climate data: http://www.cru.uea.ac.uk/cru/data/temperature/#datter
and ftp://ftp.cmdl.noaa.gov/
Gene Expression Omnibus: http://www.ncbi.nlm.nih.gov/geo/
Geo Spatial Data: http://geodacenter.asu.edu/datalist/
Human Microbiome Project: http://www.hmpdacc.org/reference_genomes/reference_genomes.php
MIT Cancer Genomics Data: http://www.broadinstitute.org/cgi-bin/cancer/datasets.cgi
NASA: http://nssdc.gsfc.nasa.gov/nssdc/obtaining_data.html
NIH Microarray data: ftp://ftp.ncbi.nih.gov/pub/geo/DATA/supplementary/series/GSE6532/ (R)
Protein structure: http://www.infobiotic.net/PSPbenchmarks/
Public Gene Data: http://www.pubgene.org/
Stanford Microarray Data: http://smd.stanford.edu//
Social Sciences
General Social Survey: http://www3.norc.org/GSS+Website/
ICPSR: http://www.icpsr.umich.edu/icpsrweb/ICPSR/access/index.jsp
UCLA Social Sciences Archive: http://dataarchives.ss.ucla.edu/Home.DataPortals.htm
UPJOHN INST: http://www.upjohn.org/erdc/erdc.html
Time Series
Time Series data Library: http://robjhyndman.com/TSDL/
Universities
Carnegie Mellon University Enron email: http://www.cs.cmu.edu/~enron/
Carnegie Mellon University StatLab: http://lib.stat.cmu.edu/datasets/
Carnegie Mellon University JASA data archive: http://lib.stat.cmu.edu/jasadata/
Ohio State University Financial data: http://fisher.osu.edu/fin/osudata.htm
UC Berkeley: http://ucdata.berkeley.edu/
UCLA: http://wiki.stat.ucla.edu/socr/index.php/SOCR_Data
UC Riverside Time Series: http://www.cs.ucr.edu/~eamonn/time_series_data/
University of Toronto: http://www.cs.toronto.edu/~delve/data/datasets.html
Here follows a list of data sources that may easily be imported into R.
If an (R) appears after source this means that the data are already in R format or there exist R commands for directly importing the data from R. (See http://www.quantmod.com/examples/intro/ for some code.) Otherwise, i have limited the list to data sources for which there is a reasonably simple process for importing csv files. What follows is a list of data sources organized into categories that are not mutually exclusive but which reflect what's out there.
Economics
UMD:: http://inforumweb.umd.edu/econdata/econdata.htmlWorld bank: http://data.worldbank.org/indicator
Finance
CBOE Futures Exchange: http://cfe.cboe.com/Data/Google Finance: http://finance.yahoo.com/ (R)
Google Trends: http://www.google.com/trends?q=google&ctab=0&geo=all&date=all&sort=0
St Louis Fed: http://research.stlouisfed.org/fred2/ (R)
NASDAQ: https://data.nasdaq.com/
OANDA: http://www.oanda.com/ (R)
Yahoo Finance: http://finance.yahoo.com/ (R)
Government
Archived national government statistics: http://www.archive-it.org/Australia: http://www.abs.gov.au/AUSSTATS/abs@.nsf/DetailsPage/3301.02009?OpenDocument
Canada: http://www.data.gc.ca/default.asp?lang=En&n=5BCD274E-1
DataMarket: http://datamarket.com/
Fed Stats: http://www.fedstats.gov/cgi-bin/A2Z.cgi
Guardian world governments: http://www.guardian.co.uk/world-government-data
London, U.K. data: http://data.london.gov.uk/catalogue
New Zealand: http://www.stats.govt.nz/tools_and_services/tools/TableBuilder/tables-by...
NYC data: http://nycplatform.socrata.com/
OECD: http://www.oecd.org/document/0,3746,en_2649_201185_46462759_1_1_1_1,00.html
San Francisco Data sets: http://datasf.org/
U.K. Government Data:http://data.gov.uk/data
United Nations: http://data.un.org/
U.S. Federal Government Agencies: http://www.data.gov/metric
US CDC Public Health datasets: http://www.cdc.gov/nchs/data_access/ftp_data.htm
The World Bank: http://wdronline.worldbank.org/
Machine Learning
Causality Workbench: http://www.causality.inf.ethz.ch/repository.phpKaggle competition data: http://www.kaggle.com/
KDNuggets competition site: www.kdnuggets.com/datasets/
UCI Machine Learning Repository: http://archive.ics.uci.edu/ml/
Machine Learning Data Set Repository: http://mldata.org/
Microsoft Research: http://research.microsoft.com/apps/dp/dl/downloads.aspx
Million songs: http://blog.echonest.com/post/3639160982/million-song-dataset
Social Networking: http://www.cs.cmu.edu/~jelsas/data/ancestry.com/
Public Domain Collections
Data360: http://www.data360.org/index.aspxDatamob.org: http://datamob.org/datasets
Factual: http://www.factual.com/topics/browse
Freebase: http://www.freebase.com/
Google: http://www.google.com/publicdata/directory
infochimps: http://www.infochimps.com/
numbray: http://numbrary.com/
Sample R data sets: http://stat.ethz.ch/R-manual/R-patched/library/datasets/html/00Index.html (R)
SourceForge Research Data: http://www.nd.edu/~oss/Data/data.html
UFO Reports: http://www.nuforc.org/webreports.html
Wikileaks 911 pager intercepts: http://911.wikileaks.org/files/index.html
Stats4Stem.org: R data sets: http://www.stats4stem.org/data-sets.html (R)
The Washington Post List: http://www.washingtonpost.com/wp-srv/metro/data/datapost.html
Science
Agricultural Experiments: http://www.inside-r.org/packages/cran/agridat/docs/agridat (R)Climate data: http://www.cru.uea.ac.uk/cru/data/temperature/#datter
and ftp://ftp.cmdl.noaa.gov/
Gene Expression Omnibus: http://www.ncbi.nlm.nih.gov/geo/
Geo Spatial Data: http://geodacenter.asu.edu/datalist/
Human Microbiome Project: http://www.hmpdacc.org/reference_genomes/reference_genomes.php
MIT Cancer Genomics Data: http://www.broadinstitute.org/cgi-bin/cancer/datasets.cgi
NASA: http://nssdc.gsfc.nasa.gov/nssdc/obtaining_data.html
NIH Microarray data: ftp://ftp.ncbi.nih.gov/pub/geo/DATA/supplementary/series/GSE6532/ (R)
Protein structure: http://www.infobiotic.net/PSPbenchmarks/
Public Gene Data: http://www.pubgene.org/
Stanford Microarray Data: http://smd.stanford.edu//
Social Sciences
General Social Survey: http://www3.norc.org/GSS+Website/ICPSR: http://www.icpsr.umich.edu/icpsrweb/ICPSR/access/index.jsp
UCLA Social Sciences Archive: http://dataarchives.ss.ucla.edu/Home.DataPortals.htm
UPJOHN INST: http://www.upjohn.org/erdc/erdc.html
Time Series
Time Series data Library: http://robjhyndman.com/TSDL/ Universities
Carnegie Mellon University Enron email: http://www.cs.cmu.edu/~enron/Carnegie Mellon University StatLab: http://lib.stat.cmu.edu/datasets/
Carnegie Mellon University JASA data archive: http://lib.stat.cmu.edu/jasadata/
Ohio State University Financial data: http://fisher.osu.edu/fin/osudata.htm
UC Berkeley: http://ucdata.berkeley.edu/
UCLA: http://wiki.stat.ucla.edu/socr/index.php/SOCR_Data
UC Riverside Time Series: http://www.cs.ucr.edu/~eamonn/time_series_data/
University of Toronto: http://www.cs.toronto.edu/~delve/data/datasets.html
china's history of 5000 years - in songs
歌诀一(大陆人教版小学历史教材)
夏商與西周,東周分兩段。
春秋與戰國,一統秦兩漢。
三分魏蜀吳,兩晉前後延。
南北朝並立,隋唐五代傳。
宋元明清後,皇朝至此完。
春秋與戰國,一統秦兩漢。
三分魏蜀吳,兩晉前後延。
南北朝並立,隋唐五代傳。
宋元明清後,皇朝至此完。
歌诀二 (達拉斯市李嘉清所紀)
唐虞夏商周完東,春秋戰國秦末烽。
漢分三國收魏晉,南北朝夕隋唐風。
五代十國遼夏匆,宋疲金盡元朝攻。
明滅清寒虛國庫,國民革命歸於中。
漢分三國收魏晉,南北朝夕隋唐風。
五代十國遼夏匆,宋疲金盡元朝攻。
明滅清寒虛國庫,國民革命歸於中。
歌诀三(大陆苏教版历史教材)
唐堯虞舜夏商周,春秋戰國亂悠悠。
秦漢三國晉統一,南朝北朝是對頭。
隋唐五代又十國,宋元明清帝王休。
秦漢三國晉統一,南朝北朝是對頭。
隋唐五代又十國,宋元明清帝王休。
歌訣四(香港中學生常用簡易版本)
黃虞夏商周,春秋戰國秦,
兩漢三國晉,晉後南北分,
隋唐五代宋,元明清及民。
兩漢三國晉,晉後南北分,
隋唐五代宋,元明清及民。
歌訣五(1972,1973 年馬來西亞古晉中學劉延森老師)
三皇五帝夏商周,戰國歸秦繼漢劉,
三國魏晉南北繼,隋唐五代宋元明。
三國魏晉南北繼,隋唐五代宋元明。
歌诀六
盤古開天神話傳, 三皇五帝數千年。
炎帝黃帝華夏祖, 堯舜禹王位讓賢。
夏商西周奴隸制, 東周列國變封建。
秦漢統一開疆域, 三國紛爭起戰亂。
西晉東晉南北朝, 隋唐疆域又擴展。
五代十國鬧割據, 宋遼夏金歸大元。
明朝船隊下西洋, 清朝鎖國被破關。
民國內戰加外戰, 人民共和開新篇。
炎帝黃帝華夏祖, 堯舜禹王位讓賢。
夏商西周奴隸制, 東周列國變封建。
秦漢統一開疆域, 三國紛爭起戰亂。
西晉東晉南北朝, 隋唐疆域又擴展。
五代十國鬧割據, 宋遼夏金歸大元。
明朝船隊下西洋, 清朝鎖國被破關。
民國內戰加外戰, 人民共和開新篇。
歌诀七
三皇五帝夏商周,歸秦及漢三國謀。
晉終南北隋唐記,五代宋元明清華。
晉終南北隋唐記,五代宋元明清華。
歌诀八
三皇五帝夏商周,春秋戰國亂悠悠。
秦漢三國傳兩晉,南朝北朝是對頭。
隋唐五代北南宋,元明清後帝王休。
秦漢三國傳兩晉,南朝北朝是對頭。
隋唐五代北南宋,元明清後帝王休。
歌诀九(中华上下五千年)
盘古开天地,
中华立根基!
三皇五帝夏商周,
春秋战国百家聚
秦王扫六合,
大汉雄风起
三国鼎立,
两晋南北朝
隋唐又统一
唐威通天下,
宋韵建海隅!
辽金元明清兴衰交替
辛亥风云急。
血肉筑长城,
古国迎晨曦!
东方醒狮,
昂首腾飞!
自强永不息。
东方醒狮,
昂首腾飞!
自强永不息!
中华立根基!
三皇五帝夏商周,
春秋战国百家聚
秦王扫六合,
大汉雄风起
三国鼎立,
两晋南北朝
隋唐又统一
唐威通天下,
宋韵建海隅!
辽金元明清兴衰交替
辛亥风云急。
血肉筑长城,
古国迎晨曦!
东方醒狮,
昂首腾飞!
自强永不息。
东方醒狮,
昂首腾飞!
自强永不息!
歌诀十
三皇五帝夏商周,春秋戰國亂悠悠。
秦漢三國晉十六,南朝北朝是對頭。
隋唐五代又十國,宋元明清帝王休。
秦漢三國晉十六,南朝北朝是對頭。
隋唐五代又十國,宋元明清帝王休。
歌诀十一
曲为《只要我長大》:唐堯虞舜夏商周
秦漢接三國
魏晉南北朝 隋唐五代十國
宋元明清 中華民國
中華文化五千年
我們要發揚
我們要光大
秦漢接三國
魏晉南北朝 隋唐五代十國
宋元明清 中華民國
中華文化五千年
我們要發揚
我們要光大
2011年12月26日星期一
see outside of you are allowed - anti-block tools for internet surfing
http://jingpin.org/100-anti-censorthip-tools/
100 个免费翻墙工具
100 个免费翻墙工具及其使用方法:
一、在线代理网站 (66)
在线代理网站的最大好处是你不需要下载安装任何的软件,最不好的地方就是弹窗广告很烦人:1、Aniscartujo.com
这个 Aniscartujo 在线代理适用于电脑和手机。
2、Free Web Proxy
Free Web Proxy 可以观看 YouTube 视频并将它们下载为 MP4 文件。
3、Daveproxy
不能看 YouTube 视频。
4、TryCatchMe
不能看 YouTube 视频。
5、Surfagain.com
Surfagain.com 可以看 YouTube 视频。
6、Online Sonic
Online Sonic 会将你访问的网站直接转换成法语版本。
7、Megaproxy
免费的 Magaproxy 代理没有弹窗广告。
8、Shield Proxy
这个网站设计简单,主页基本上就一个大的输入框。
9、Psiphon 2
Psiphon 2 需要邀请才能使用,并且网站地址经常更改。
10、Glype
你可以通过 Glype 创建自己的在线代理网站,只需要把它的脚本上传到服务器。
11、Circumventor
Circumventor 会在网站上提供一个在线代理的链接,如果该链接对应的网站被墙,你可以输入邮箱地址并订阅它的其他在线代理。
12-47、CamoList.com
CamoList.com 网站有提供 30 多个在线代理,并且全部都可以用来看 YouTube 视频:
- artclassdrama.com
- browse007.com
- browse007.info
- camo1.info
- classwork101.com
- coolkidsonly.org
- ditchthetests.com
- downwithitall.com
- dumbdream.com
- enoughschool.com
- erasermaker.com
- forgotmybooks.com
- getus.in
- goodgradesforme.com
- gumunderthedesk.com
- gymtimestories.com
- hiddentunnel.net
- hidemy.biz
- letmethruthis.com
- nobodycanstop.us
- noclasswork.com
- noneedhallpass.com
- nowaytoknow.com
- plzhidemy.info
- rebelbrowse.com
- schoolisgood.com
- showsomewisdom.com
- slaptheblock.com
- sneakmyass.in
- starscantshine.com
- studybreakneeded.com
- studyhardplayharder.com
- theunblocked.com
- tothedeans.com
- tunnel007.com
- wecantfocus.com
Polysolve.com 网站有提供 18 个在线代理的链接(包括它自己的):
- Atunnel.com
- Backfox.com
- Btunnel.com
- Calculatepie.com
- Ctunnel.com
- Dtunnel.com
- Englishtunnel.com
- Geotunnel.com
- Mathtunnel.com
- Newbackdoor.com
- Polysolve.com
- SafeForWork.net
- Safehazard.com
- Safelizard.com
- USAtunnel.com
- Vmathpie.com
- VPNTunnel.net
- Vtunnel.com
66、Proxy.org (100+)
Proxy.org 网站有提供 100 多个可用的免费在线代理。
二、客户端代理 (11)
客户端代理基本上都是没有商业广告的,只是需要现在安装才能使用:- 自由门(适用于 Windows、 Windows Mobile 以及一些 Java 手机)
- 逍遥游(类似自由门,由同一网站提供)
- 动网通(类似自由门,由同一网站提供)
- 无界 (适用于 Windows)
- GTunnel (适用于 Windows)
- Tor (适用于 Windows、Mac、Linux、Android、iOS 以及 Nokia)
- GappProxy(适用于 Windows 和 Linux)
- Hyk-proxy (适用于 Windows、Linux 和 Mac)
- Your Freedom (适用于 Windows 和 Mac)
- GPass (适用于 Windows)
- HTTP-Tunnel (适用于 Windows)
在使用以上 11 种代理软件的时候,你需要修改浏览器网络的代理地址,而以下扩展可以让修改的过程变得简单,并且可以只对被墙的网站起作用:
- AutoProxy (适用于 Firefox)
- Proxy Switchy (适用于 Chrome)

三、VPN (13)
代理工具只是对那些设置了网络连接方式的应用程序生效,并且绝大多数只适用于电脑,而 VPN (Virtual Private Network) 则是对所有程序生效,并且大多数适用于电脑和手机。a、免费免安装的 VPN (6)
绝大多数不需要安装就可以使用的 VPN 都适用于任何系统的电脑和智能手机:
- Tsunagarumon
- MacroVPN
- USA IP
- Prox Network
- Free UK VPN
- Best Free VPN
b、免费 VPN 客户端 (7)
需要安装的 VPN 客服端大多只适用于电脑:
- SecurityKiss (适用于 Windows)
- ProXPN (适用于 Windows 和 Mac)
- Hotspot Shield (适用于 Windows 和 Mac)
- ExpatShield (适用于 Windows)
- Loki VPN Client (适用于 Windows)
- Free OpenVPN Service (适用于 Windows、Mac、Linux、iOs、Android、等等)
- RaptorVPN (适用于 Windows 和 Mac)
四、SSH (3)
如之前所说,免费的 SSH 并不稳定,但是你仍然可以尝试以下三个:以上三个 SSH 服务都是中文的。
备注:
获得 SSH 帐号之后,你还需要可以启用该帐号的工具,例如适用于 Windows 的 Tunnelier,以及适用于 Mac 的 Issh。
五、浏览器 (3)
最优秀的浏览器应该可以直接翻墙。a、电脑浏览器(1)
Firefox、Chrome、IE 以及其他流行的浏览器都不可以直接翻墙,但是 Alkasir 可以。
b、手机浏览器(2)
手机可以通过以下两个浏览器直接翻墙:
不过可惜的是以上两个浏览器在国内都被和谐了,孟加拉、伊朗、越南等国的朋友可能还用得上。
六、IPv6 (1)
如果你的宽带(例如教育网的)支持 IPv6,那么就可以直接访问那些有 IPv6 地址的被墙网站;否则,你也可以利用一些第三方工具(例如 gogoCLIENT)访问那些网站。下载并安装好 gogoCLIENT 软件之后,你就可以通过以下三种方法访问 Google、Twitter、YouTube 以及其他支持 IPv6 的被墙网站:
1、在网站域名后面添加 .sixxs.org 后缀,例如:通过 Google.com.sixxs.org 访问 Google 网站;
2、使用自动代理服务,即 “proxy.pac” 链接,例如:
http://gfw-proxy.co.cc/proxy.pac3、在 hosts 文件里面添加被墙网站的 IPv6 地址。其中 Windows 用户可以在以下路径找到该文件:
C:\WINDOWS\system32\drivers\etc另外,你可以在这个 Google 文档 上找到 Google、YouTube、Twitter 等多个流行被墙网站的 IPv6 地址。
备注:
如果没有 IPv6 网络,你也可以在 hosts 文件里面添加那些被墙网站的尚未被墙的 IPv4 地址 (即通常所说的 IP),然后就可以不需要任何的工具直接访问这些网站。另外,你可以通过 OpenDNS 网站轻松地找到某个域名的所有 IPv4 地址。
七、其他 (3)
以上所提到的工具都是用来直接突破网络封锁的,除此之外,你也可以通过以下 3 种方法间接翻墙:98. Google 网页缓存
如果某个被墙的网站有被 Google 搜录,那么你就可以通过 Google 搜索结果的网页缓存查看该网站。
99、Google 翻译
通过 Google 翻译,你只要输入被墙网页的链接,并选择一种不同的语言,然后你就可以浏览该网页的译文,甚至原文。
100、Google 阅读器
现在很多网站都有提供 RSS Feed,所以你可以通过 Google 阅读器订阅它们。
关于以上三种间接翻墙方法的不足和更详细介绍,可以参考《如何翻墙》一文。
备注
在以上 100 个免费的翻墙工具里面,很难说哪个最好,因为每一个都随时有可能被墙,所以多一个就多一份保障。Research Blogging Awards 2010
The Winners
Finalists for each award are listed in random order.
- Research Blog of the Year $1,000
Winner: Not Exactly Rocket Science (RB page)
Finalists:
明夷待访录 - Chinese book of Du Contrat Social
http://zh.wikisource.org/wiki/%E6%98%8E%E5%A4%B7%E5%BE%85%E8%A8%AA%E9%8C%84
http://book.douban.com/subject/1766308/
http://book.douban.com/subject/1766308/
50 botanical blogs
http://www.onlinecollegecourses.com/2011/02/16/50-best-blogs-for-botany-students/
Botanical Blogs
These blogs are botany-focused, providing news and information you can use as a student and future plant scientist.
Use these blogs to learn about some new plant genera and species, some of which are amazingly unique.
Hear from professors, botanists and a wide range of plant experts on these great blogs.
These students share their research and passion for plants though their blogs.
If you want a blog that focuses on one type of plant, ecosystem or botany subject, these sites are excellent resources.
Botanic gardens are great places to learn more about plants and to see some rare and beautiful varieties in person. If you don’t live near one or just want a quick botanical fix, visit these blogs instead.
Plants can be wonderful on their own, but much human interaction with plants has to do with using and manipulating them to suit our own needs. That’s where these blogs come in. They’ll teach you the essentials of raising plants for pleasure and for sustenance.
Botanical Blogs
These blogs are botany-focused, providing news and information you can use as a student and future plant scientist.
- AoB Blog: If you’re planning on being a professional in the field of botany, bookmark this blog now. You’ll get updates about the latest research in the field from the Annals of Botany.
- Plant Science Blog: A division of the Biology Blog, this site offers up some of the latest discoveries in plant science, with new posts regularly.
- Plants and Botany: This group of plant lovers post pictures, questions and discussions on this blog, which can be a great way for newbies to the field to learn more.
- Foothills Fancies: Embrace the wonders of the natural world with this blog, written by naturalist S.L. White.
- PAPGREN: The Pacific Agricultural Genetic Resource Network supplies those interested in plant genetic research and discovers with news and helpful articles here.
- Berry Go Round: Looking for the best posts and blogs to read about plants? This blog hosts a monthly carnival, with links to a wide range of interesting botanical material.
- Blog: Botany: On this blog you’ll find posts from Dr. Robson of The Manitoba Museum. She shares some interesting stories about the plants the museum houses.
- Thomas’ Plant-Related Blog: Blogger Thomas loves plants, and on this site you’ll find numerous articles on interesting plant species, from plankton to broccoli.
Use these blogs to learn about some new plant genera and species, some of which are amazingly unique.
- Botany Blog: This blog is a must-read for any botany student or plant lover. Readers will find regular posts highlighting one particular plant species– a great way to learn more about the innumerable plant species out there.
- Net World Directory Botany Blog: Check back with this blog often to learn more about specific plants as well as their cultivation and use.
- Get Your Botany On!: This blog is filled with beautiful images and descriptions of plants as well as some posts touching on important news items in the world of botany.
- Plants are the Strangest People: Here you’ll find a plant-loving blogger who works in a garden center, posting on everything from useful plant resources to the care and feeding of houseplants.
- Exploring the World of Trees: Want to know more about the tree species of the world? This blog is a good place to start, with posts focusing on specific types of trees accompanied by pretty pictures of them.
Hear from professors, botanists and a wide range of plant experts on these great blogs.
- Niches: Here, a Georgia bloggers and plant biologists Wayne and Glenn discuss native plants, habitats and the field at large.
- Talking Plants: Jaime Plaza of the Sydney Botanic Gardens shares photos and information on plants on this blog.
- The Phytophactor: This blogger shares a love of plants on this site, but also works as a botanist focusing on economic botany, rain forest ecology and plant diversity.
- Biofortified: With a wide range of professionals in plant science fields posting to this site, it’s a great read for anyone interested in the genetic manipulation of plants for agricultural purposes.
- Invasive Species Weblog: Jennifer Forman Orth, an invasive plant ecologist, shares her expertise on invasive plant problems troubling biologists the world over.
- California Botany Blog: Dean William Taylor offers a look at some of his research into flowers and seeds on this blog.
- Seeds Aside: Focusing on plant evolution and ecology, this blogger shares what he’s learning in botany with readers on this site.
- Botanizing: Larry Hufford, Professor of Biology, posts about his expeditions into the natural world here, with lovely photos to illustrate.
- A Digital Botanic Garden: This blog is the home of Phil Gates, a botanist working at Durham University. You’ll find excellent posts on a wide variety of plant life that will help you learn about and marvel at the plant world.
These students share their research and passion for plants though their blogs.
- My Growing Passion: Margaret Morgan is studying to get her degree in Biology, with a focus on plant life, but she also just plain has a passion for botanicals. On this site, see posts that reflect both her interest in growing and learning about plants.
- James and the Giant Corn: Grad and doctoral students in botany can take a peek into another student’s work through this blog from Berkeley student James.
- Moss Plants and More: Read through this blog to learn more about bryology, the study of mosses, from graduate student Jessica.
If you want a blog that focuses on one type of plant, ecosystem or botany subject, these sites are excellent resources.
- Cactus Blog: If you prefer your plants pointy and leafless, then check out this cactus-centric blog. You’ll learn about all things succulent and cacti related.
- Wild Plants Post: While cultivated plants can be great, this blog chooses to focus on their wild cousins, sharing posts about ecosystems and evolution as well.
- My Orchids Journal: Many houseplant enthusiasts love to grow orchids and other tropical flowering plants. Learn more about what it takes to get them growing right from this blog.
- Early Forest: Those with a passion for trees and forestry will find this blog, and its amazing photos, a great inspiration.
- Treeblog: Can’t get enough of those monsters of the plant world, trees? This blog is full of great information and guidance on how to learn more.
- No Seeds, No Fruits, No Flowers: No Problem: With hundreds of different and diverse varieties, ferns have fascinated people for hundreds of years. Learn more about these beautiful plants through this blog.
- SwampThings: You’ll get a chance to better understand the ecology of the plants and animals that call the swamp home when you read this blog.
- The Plant Mafia Blog: These plant lovers are committed to raising beautiful, healthy plants and share them with each other, readers and local botanical gardens.
Botanic gardens are great places to learn more about plants and to see some rare and beautiful varieties in person. If you don’t live near one or just want a quick botanical fix, visit these blogs instead.
- Botany Photo of the Day: The UBC Botanical Garden posts a new photo of the gorgeous plants they care for on this blog every few days.
- Plant Talk: This blog from the New York Botanical Garden is a great source of information not only about the gardens but about plant life in general.
- Denver Botanic Gardens: Find out just what’s happening in Denver’s Botanic Gardens from this blog.
- Lewis Ginter Botanical Garden: See photos of the beautiful greenhouses and gardens at this botanical garden on their blog, and get advice on great books, plant care and much more.
- Ogden Botanical Gardens: This blog will inspire your love of plants even more, with information about classes, seasonal bloomers and gardening activities.
- The Dig!: Green Bay Botanical Gardens share just what’s happening every season of the year, letting those in colder climes know when its worth it to brave the weather to see beautiful plants.
- Norfolk Botanical Garden: With detailed posts about the trees and flowers in their collection, this blog is a great read whether you can visit these gardens or not.
Plants can be wonderful on their own, but much human interaction with plants has to do with using and manipulating them to suit our own needs. That’s where these blogs come in. They’ll teach you the essentials of raising plants for pleasure and for sustenance.
- Garden Voices: If you don’t have time to browse multiple blogs, consider this blog that collects some of the best gardening posts from the web.
- Garden Rant: You’ll find gardening advice aplenty on this excellent horticultural blog.
- Agricultural Biodiversity Weblog: Read through this blog to gain a better understanding of what agricultural biodiversity is so important to world food supplies.
- Landscape Juice: With posts on gardening, landscape ecology, tree planting and other plant-focused matters, this blog is an excellent resource for those interested in botany or horticulture.
- Heavy Petal: Learn more about organic, urban gardening from this great blog.
- In the Herb Garden: The Herb Companion Magazine is home to this blog containing posts from a number of gardeners and herb lovers.
- Growing With Plants: If you want help deciding what to plant in your own backyard or just want to learn more about decorative plants, check out this blog.
- The Plant Hunter: Tim Wood travels the world in search of the coolest plants for home and garden and posts about them here.
- Love Plant Life Blog: This blog is focused on agriculture and growing food and while you can learn more about plants, you’re likely to learn more about policy.
- Plant Guides Blog: Need access to a helpful plant growing guide? This site is full of them.
2011年12月25日星期日
what is remainder or modulus in python
http://stackoverflow.com/questions/509710/python-quotient-vs-remainder
Modulo is performed in the integer context, not fractional (remainders are integers). Therefore:
Modulo is performed in the integer context, not fractional (remainders are integers). Therefore:
1 % 1 = 0 (1 times 1 plus 0)
1 % 2 = 1 (2 times 0 plus 1)
1 % 3 = 1 (3 times 0 plus 1)
6 % 3 = 0 (3 times 2 plus 0)
7 % 3 = 1 (3 times 2 plus 1)
8 % 3 = 2 (3 times 2 plus 2)
Please give credit where credit is due
Please give credit where credit is due. Plant biologists would rightfully be ridiculed if they claimed to have made new discoveries while equivalent phenomena were already known from animals or fungi. Given that the value of the world's agriculture is more than three times that of the entire pharmaceutical industry and that many more people die each year of hunger and malnutrition than from cancer, it is time that scientists of all stripes paid more attention to plant biology.
http://www.cell.com/fulltext/S0092-8674%2811%2901501-7
http://www.cell.com/fulltext/S0092-8674%2811%2901501-7
2011年12月24日星期六
regular expressions and R
http://r-ecology.blogspot.com/2011/10/r-tutorial-on-regular-expressions-regex.html
visualzing/graphical tools/books
(1) protovis, D3.js - A graphical toolkit for visualization
http://mbostock.github.com/protovis/ex/
(2) Beautiful visualization
http://www.amazon.com/Beautiful-Visualization-Looking-through-Practice/dp/1449379869
(3) Data flow
http://www.amazon.com/Data-Flow-Visualising-Information-Graphic/dp/3899552172/ref=pd_sim_b_6
(4)Visualize this
http://www.amazon.com/Visualize-This-FlowingData-Visualization-Statistics/dp/0470944889/ref=pd_sim_b_1
(5) Beautiful data
http://www.amazon.com/Beautiful-Data-Stories-Elegant-Solutions/dp/0596157118/ref=pd_sim_b_4
http://mbostock.github.com/protovis/ex/
(2) Beautiful visualization
http://www.amazon.com/Beautiful-Visualization-Looking-through-Practice/dp/1449379869
(3) Data flow
http://www.amazon.com/Data-Flow-Visualising-Information-Graphic/dp/3899552172/ref=pd_sim_b_6
(4)Visualize this
http://www.amazon.com/Visualize-This-FlowingData-Visualization-Statistics/dp/0470944889/ref=pd_sim_b_1
(5) Beautiful data
http://www.amazon.com/Beautiful-Data-Stories-Elegant-Solutions/dp/0596157118/ref=pd_sim_b_4
2011年12月23日星期五
Software Carpentry - Helping scientists make better software/programming
http://software-carpentry.org/
Version 4
We are currently updating the content and format so that students can work through the material they need, when they need it. The lectures we have completed so far cover:- Version Control – Learn how to collaborate with other people and automatically create a record of previous work using a version control system.
- The Shell – Much of scientific computing involves the Unix operating system. Effectively using the shell is one of the first steps to efficient Unix programming.
- Python – A versatile open source language that is increasingly popular among scientific programmers.
- Testing – The basics of software testing, including exception handling and unit testing.
- Sets and Dictionaries – Using associative data structures to better represent data that isn’t a list or vector.
- Regular Expressions – Manipulate text quickly with this powerful set of pattern matching tools.
- Databases – An introduction to SQL, the most popular database query language.
- Classes and Objects – The basics of object-oriented programming.
- Program Design – An example driven introduction to effective program design.
- Systems Programming – How to manipulate files and directories from a program.
- Make – This tool will help automate everything from large software builds to batch processes.
- Matrix Programming – Use array libraries to make numerical programs smaller and faster.
- MATLAB – The world’s most popular numerical programming language.
- Multimedia Programming – Work with images, sound, and other media.
- Spreadsheets – Learn to use spreadsheets for data organization, analysis, and visualization.
- Essays – Longer (non-video) discussion of some important ideas in scientific programming.
- Recommended Reading – An annotated bibliography.
- Glossary – Key terms.
Version 3
The lecture notes from Version 3 of this course (2004-2009) are available at http://software-carpentry.org/3_0/.general advice to undergraduates who are interested in science
http://blog.ketyov.com/2011/08/career-advice.html
Here is a blog article, listed some advice to undergraduates.
It took me a while to respond for several reasons, not the least of which is that I'm super busy. And also because it felt kind of weird to answer. I mean, I didn't study neuroscience as an undergrad. And I didn't do well as an undergrad. So I certainly shouldn't be doling out advice!
(1) It's important to me that other people know how hard this life, science, and career stuff really is. People should know that often, success doesn't come easy.
(2) The second piece of non-specific advice: learn to network. Talk to other researchers. Email people about their work when you have questions. Don't be shy. Or rather, go ahead and be shy but recognize that lots of people are shy and the only way to learn from them is to overcome your mutual shyness. Plus, researchers love to know that someone read their work and are interested.
This advice isn't meant as a machiavellian ploy or anything. Networking lets you meet smart people, which gives you new ideas and new collaborations. This, in turn, lets you do science faster and better.
Networking is sharing, not manipulating.
(3) Third: learn how to do your own data analysis. Know statistics well. Know at least some basic programming/scripting in Python, R, Matlab, etc. This will be of immense value in helping you get your research done efficiently and correctly, without needing to rely on other people's code (and time and commitment). This will become more important as our field becomes more data driven.
Here is a blog article, listed some advice to undergraduates.
It took me a while to respond for several reasons, not the least of which is that I'm super busy. And also because it felt kind of weird to answer. I mean, I didn't study neuroscience as an undergrad. And I didn't do well as an undergrad. So I certainly shouldn't be doling out advice!
(1) It's important to me that other people know how hard this life, science, and career stuff really is. People should know that often, success doesn't come easy.
(2) The second piece of non-specific advice: learn to network. Talk to other researchers. Email people about their work when you have questions. Don't be shy. Or rather, go ahead and be shy but recognize that lots of people are shy and the only way to learn from them is to overcome your mutual shyness. Plus, researchers love to know that someone read their work and are interested.
This advice isn't meant as a machiavellian ploy or anything. Networking lets you meet smart people, which gives you new ideas and new collaborations. This, in turn, lets you do science faster and better.
Networking is sharing, not manipulating.
(3) Third: learn how to do your own data analysis. Know statistics well. Know at least some basic programming/scripting in Python, R, Matlab, etc. This will be of immense value in helping you get your research done efficiently and correctly, without needing to rely on other people's code (and time and commitment). This will become more important as our field becomes more data driven.
Markov Chain - interesting and attractive
http://freakonometrics.blog.free.fr/index.php?post/2011/12/20/Basic-on-Markov-Chain-%28for-parents%29
2011年12月22日星期四
PGDD - plant genome duplication database
http://chibba.agtec.uga.edu/duplication/index/home
PGDD is a public database to identify and catalog plant genes in terms of intragenome or cross-genome syntenic relationships. Current efforts focus on flowering plants with available whole genome sequences (preferrably assembled pseudomolecules with ordered gene models).
PGDD is a public database to identify and catalog plant genes in terms of intragenome or cross-genome syntenic relationships. Current efforts focus on flowering plants with available whole genome sequences (preferrably assembled pseudomolecules with ordered gene models).
guide for genotype imputation tools - imput2
https://mathgen.stats.ox.ac.uk/impute/impute_v2.html#program_options
estimate or interpolate recombination rate for you
http://pbil.univ-lyon1.fr/software/mareymap/index.php
MareyMap is a meiotic recombination rate estimation program. It is based on R and features a graphical interfcace in tcl/tk. MareyMap comes with an extensive dataset and several interpolation methods. The user may also use MareyMap with his own data. A more detailed description of the capabilities of the program can be found in the section features .
from Genetic map to genetic distance among physical markers. Especially for Arabidopsis thaliana.
MareyMap is a meiotic recombination rate estimation program. It is based on R and features a graphical interfcace in tcl/tk. MareyMap comes with an extensive dataset and several interpolation methods. The user may also use MareyMap with his own data. A more detailed description of the capabilities of the program can be found in the section features .
from Genetic map to genetic distance among physical markers. Especially for Arabidopsis thaliana.
2011年12月20日星期二
Special Issue: Collaborative Bioinformatics and RNA Analysis
http://bib.oxfordjournals.org/content/current
- Paolo Romano,
- Rosalba Giugno,
- and Alfredo Pulvirenti
Tools and collaborative environments for bioinformatics research
Brief Bioinform (2011) 12(6): 549-561 doi:10.1093/bib/bbr055-
- Andrea Splendiani,
- Michaela Gündel,
- Jonathan M. Austyn,
- Duccio Cavalieri,
- Ciro Scognamiglio,
- and Marco Brandizi
Knowledge sharing and collaboration in translational research, and the DC-THERA Directory
Brief Bioinform (2011) 12(6): 562-575 doi:10.1093/bib/bbr051 -
- Ismael Navas-Delgado,
- Alejandro Real-Chicharro,
- Miguel Ángel Medina,
- Francisca Sánchez-Jiménez,
- and José F. Aldana-Montes
Social pathway annotation: extensions of the systems biology metabolic modelling assistant
Brief Bioinform (2011) 12(6): 576-587 doi:10.1093/bib/bbq061 -
- Dario Corrada,
- Federica Viti,
- Ivan Merelli,
- Cristina Battaglia,
- and Luciano Milanesi
myMIR: a genome-wide microRNA targets identification and annotation tool
Brief Bioinform (2011) 12(6): 588-600 doi:10.1093/bib/bbr062 -
- Magdalena Rother,
- Kristian Rother,
- Tomasz Puton,
- and Janusz M. Bujnicki
RNA tertiary structure prediction with ModeRNA
Brief Bioinform (2011) 12(6): 601-613 doi:10.1093/bib/bbr050 -
- Paolo Ribeca and
- Gabriel Valiente
Computational challenges of sequence classification in microbiomic data
Brief Bioinform (2011) 12(6): 614-625 doi:10.1093/bib/bbr019
put your running accessions under nohub (running in background)
(1) put your running accessions under nohub (running in background)
http://stackoverflow.com/questions/625409/how-do-i-put-an-already-running-process-under-nohup
Using the Job Control of bash to send the process into the background:
http://stackoverflow.com/questions/625409/how-do-i-put-an-already-running-process-under-nohup
Using the Job Control of bash to send the process into the background:
> [crtl]+z
> bg
disown -h
(2) running your accessions under nohub directly
$ nohup yourcommand yourArguement &
$ nohup yourcommand yourArguement > standoutput &$ nohup yourcommandinfile.sh &
The Plant Genome: An Evolutionary View on Structure and Function
This is a special issue of the plant journal:
http://onlinelibrary.wiley.com/doi/10.1111/tpj.2011.66.issue-1/issuetoc
2011年12月13日星期二
2011年12月9日星期五
chropainter and finestructure
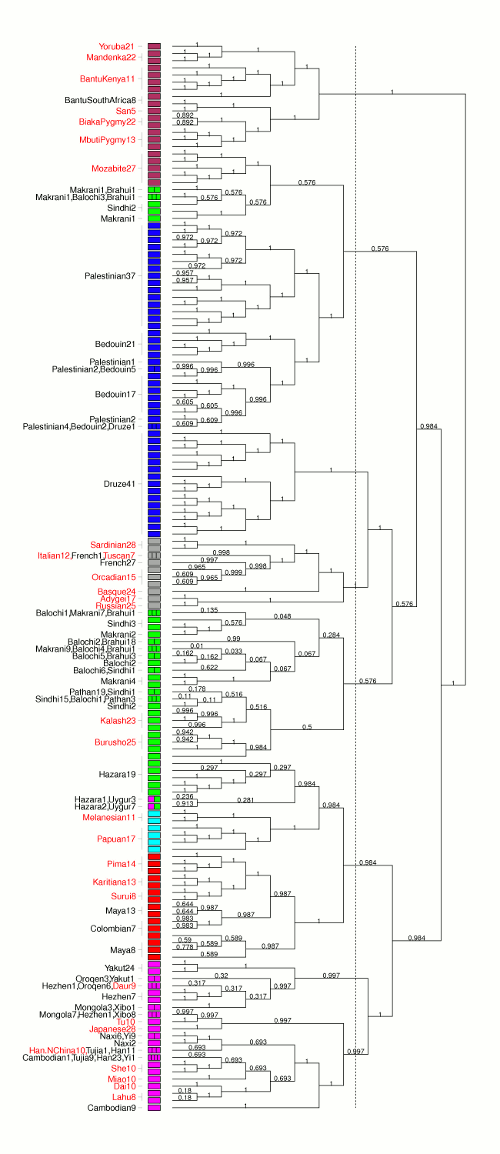
1. ChromoPainter,
http://www.paintmychromosomes.com/
ChromoPainter is a tool for finding haplotypes in sequence data. Each individual is "painted" as a combination of all other sequences. It can output a range of features, including:
2. fineSTRUCTURE is a fast and powerful algorithm for identifying population structure using dense sequencing data. By using the output of ChromoPainter as a (nearly) sufficient summary statistic, it is able to perform model-based Bayesian clustering on large datasets, including full resequencing data, and can handle up to 1000s of individuals. Full assignment uncertainty is given.
http://www.paintmychromosomes.com/
ChromoPainter is a tool for finding haplotypes in sequence data. Each individual is "painted" as a combination of all other sequences. It can output a range of features, including:
- Sample haplotypes
- Expectations of the number of recombination events at all sites
- A wide range of related features

2. fineSTRUCTURE is a fast and powerful algorithm for identifying population structure using dense sequencing data. By using the output of ChromoPainter as a (nearly) sufficient summary statistic, it is able to perform model-based Bayesian clustering on large datasets, including full resequencing data, and can handle up to 1000s of individuals. Full assignment uncertainty is given.
2011年12月2日星期五
infer population structure using dense haplotype data
It can take place of STRUCTURE, especially for whole-genome re-sequencing projects.
the tools:
http://www.maths.bris.ac.uk/~madjl/finestructure/finestructure_info.html
see this blog:
http://dienekes.blogspot.com/2011/11/chromopainter-and-finestructure.html
the tools:
http://www.maths.bris.ac.uk/~madjl/finestructure/finestructure_info.html
see this blog:
http://dienekes.blogspot.com/2011/11/chromopainter-and-finestructure.html
2011年11月18日星期五
植物碱基替换速率变异的模式和原因
The Patterns and Causes of Variation in Plant Nucleotide Substitution Rates
http://www.annualreviews.org/doi/abs/10.1146/annurev-ecolsys-102710-145119
种间杂种不亲和的遗传学
The Genetics of Hybrid Incompatibilities
http://www.annualreviews.org/doi/abs/10.1146/annurev-genet-110410-132514
这篇综述给出了种间杂种不亲和的研究方法、可能机制。
2011年11月16日星期三
2011年11月8日星期二
outcrossing and selfing in plants - evolutionary and genomic consequence
- Boris Igic, Russell Lande, Joshua R. KohnDOI: 10.1086/523362Stable URL: http://www.jstor.org/stable/10.1086/523362
 Stephen I. Wright, Rob W. Ness, John Paul Foxe, Spencer C. H. BarrettDOI: 10.1086/523366Stable URL: http://www.jstor.org/stable/10.1086/523366
Stephen I. Wright, Rob W. Ness, John Paul Foxe, Spencer C. H. BarrettDOI: 10.1086/523366Stable URL: http://www.jstor.org/stable/10.1086/523366
2011年11月6日星期日
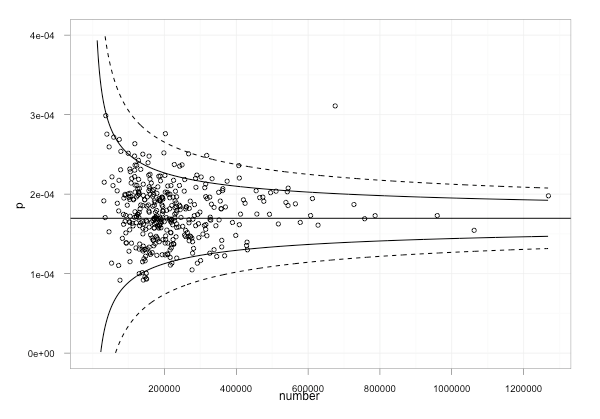
funnel plot
A funnel plot is a scatterplot of treatment effect against a measure of study size. It is used primarily as a visual aid to detecting bias or systematic heterogeneity. A symmetric inverted funnel shape arises from a ‘well-behaved’ data set, in which publication bias is unlikely. An asymmetric funnel indicates a relationship between treatment effect and study size.
A funnel plot is a useful graph designed to check the existence of publication bias in systematic reviews and meta-analyses. It assumes that the largest studies will be near the average, and small studies will be spread on both sides of the average. Variation from this assumption can indicate publication bias.
http://en.wikipedia.org/wiki/Funnel_plot
http://ouseful.files.wordpress.com/2011/10/cancerdatafunnelplot.png?w=600&h=419
A funnel plot is a useful graph designed to check the existence of publication bias in systematic reviews and meta-analyses. It assumes that the largest studies will be near the average, and small studies will be spread on both sides of the average. Variation from this assumption can indicate publication bias.
http://en.wikipedia.org/wiki/Funnel_plot
http://ouseful.files.wordpress.com/2011/10/cancerdatafunnelplot.png?w=600&h=419
2011年11月3日星期四
constrained elements in the human genome as revealed by mammalian alignments of 29 genomes
http://www.nature.com/nrg/journal/vaop/ncurrent/full/nrg3112.html
- Lindblad-Toh, K. et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 12 Oct 2011 (doi: 10.1038/nature10530)
- Lin, M. F. et al. Locating protein-coding sequences under selection for additional, overlapping functions in 29 mammalian genomes. Genome Res. 12 Oct 2011 (doi: 10.1101/gr.108753.110)
2011年10月28日星期五
What do we need to know about speciation
http://www.sciencedirect.com/science/article/pii/S0169534711002618#sec2.1
Article Outline
- The future focus of speciation research
- The origin and build-up of reproductive isolation
- Question 1. Which barriers contribute to reproductive isolation?
- Question 2. When does drift have a significant role?
- Question 3. What are the relative roles of natural and sexual selection?
- Question 4. What is the role of reinforcement?
- Question 5. How important is hybridisation in speciation?
- Question 6. What are the environmental and genetic conditions that promote speciation?
- Genetic and genomic signatures of speciation
- Connecting speciation and biodiversity
- Concluding remarks
- Online discussion forum
- Acknowledgements
- Appendix A. Supplementary data
- References
- Glossary
Quantifying effects of environmental and geographical factors on patterns of genetic differentiation
The authors used a regression-based approach to simultaneously estimate the quantitative contributions of environmental adaptation and isolation by distance on genetic variation in Boechera stricta, a wild relative of Arabidopsis.
http://onlinelibrary.wiley.com/doi/10.1111/j.1365-294X.2011.05310.x/abstract
http://onlinelibrary.wiley.com/doi/10.1111/j.1365-294X.2011.05310.x/abstract
2011年10月27日星期四
Dated origin of Arabidopsis thaliana - 13 Mya
This paper gave much more robust dating for origin of Arabidopsis thaliana:
Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana
http://www.pnas.org/content/107/43/18724.long
We bring previously overlooked fossil evidence to bear on these questions and find the split between A. thaliana and Arabidopsis lyrata occurred about 13 Mya, and that Arabidopsis and the Brassica complex (broccoli, cabbage, canola) occurred about 43 Mya.
正态检验的用途 - 似乎不大 - 要小心
1. Normality tests don't do what most think they do. Shapiro's test, Anderson Darling, and others are null hypothesis tests AGAINST the the assumption of normality. These should not be used to determine whether to use normal theory statistical procedures. In fact they are of virtually no value to the data analyst. Under what conditions are we interested in rejecting the null hypothesis that the data are normally distributed? I have never come across a situation where a normal test is the right thing to do. When the sample size is small, even big departures from normality are not detected, and when your sample size is large, even the smallest deviation from normality will lead to a rejected null.
http://stackoverflow.com/questions/7781798/seeing-if-data-is-normally-distributed-in-r/
2. I, personally, have never come across a situation where a normal test is the right thing to do. The problem is that when the sample size is small, even big departures from normality are not detected, and when your sample size is large, even the smallest deviation from normality will lead to a rejected null.
http://blog.fellstat.com/
http://stackoverflow.com/questions/7781798/seeing-if-data-is-normally-distributed-in-r/
2. I, personally, have never come across a situation where a normal test is the right thing to do. The problem is that when the sample size is small, even big departures from normality are not detected, and when your sample size is large, even the smallest deviation from normality will lead to a rejected null.
http://blog.fellstat.com/
HyPhy - Hypothesis testing using phylogenies
1. HyPhy is a scriptable package that can fit statistical evolutionary models to alignment of homologous sequences using Maximum likelihood 2), estimate various parameters that have biological meaning, for example branch lengths, substitution rates, dN/dS ratios, recombination breakpoints, and test hypotheses about how sequences in the alignment have evolved. HyPhy focuses on inference about the evolutionary process. Even though it can do limited alignment and phylogenetic reconstruction, much better specialized programs exist for these purposes.
Here are some of the applications that HyPhy is often used for:
2. Some of the most popular HyPhy functions (recombination, positive selection detection, etc) are implemented in a web-server hosted at http://www.datamonkey.org
Here are some of the applications that HyPhy is often used for:
- Positive and negative selection detection
- Recombination analysis
- Detecting co-evolving residues
- Genomic and multiple-gene evolutionary inference
- Molecular clock and relative rate tests
- Nucleotide, protein and codon model selection
- As a likelihood analysis engine for other software and web services
- One-off analyses: tasks that no other package does out of the box and are not worth writing a specialized program for
2. Some of the most popular HyPhy functions (recombination, positive selection detection, etc) are implemented in a web-server hosted at http://www.datamonkey.org
- Which codon sites are under diversifying positive or negative selection?
- Three different codon-based maximum likelihood methods, SLAC, FEL and REL, can be used estimate the dN/dS (also known as Ka/Ks or ω) ratio at every codon in the alignment. An exhaustive discussion of each approach can be found in the methodology paper. All methods can also take recombination into account. This is done by screening the sequences for recombination breakpoints, identifying non-recombinant regions and allowing each to have its own phylogentic tree.
- Is there evidence of selection in my alignment?
- The PARRIS method, developed by Konrad Scheffler and colleagues, extends traditional codon-based likelihood ratio tests to detect if a proportion of sites in the alignment evolve with dN/dS>1. The method takes recombination and synonymous rate variation into account.
- What is the evolutionary fingerprint of a gene?
- The ESD method, described in a recent paper, fits a versatile general discrete bivariate model of site-by-site selective force variation to partition all sites into selective classes, and obtains an approximate posterior distribution of this partititoning. The resulting "noisy" distribution of selective regimes is the evolutionary fingerprint of a gene. The EVF (evolutionary fingerprinting) module implements this procedure, and can also infer which individual sites appear to be positively selected while accounting for parameter estimation error (analogous to the BEB methodology of the PAML package).
- Which codon sites are under positive or negative selection at the population level?
- The codon-based maximum likelihood IFEL method can investigate whether sequences sampled from a population (e.g. viral sequences from different hosts) have been subject to selective pressure at the population level (i.e. along internal branches). A discussion of the method and its application can be found here
- Did selective pressure vary along lineages, i.e. over time?
- The codon-based genetic algorithm GABranch method can automatically partition all branches of the phylogeny describing non-recombinant data into groups according to dN/dS. Robust multi-model inference is used to collate results from all models examined during the run to provide confidence intervals on dN/dS for each branch and guard against model misspecification and overfitting (method details).
- How about episodic diversifying selection (branch-site methods)? Using the modeling framework, which allows the efficient estimations with models which permit dN/dS variation along both sites and lineages, Datamonkey implements two tests geared towards finding lineages and sites subject to episodic diversifying selection (EDS).
- The Branch-site REL method, identifies those branches where a proportion of sites evolves under EDS. If you are primarily interested in finding which lineages (but don't care about which sites) have experienced EDS, use this method. Alternatively, if you are interested in sites (but don't care about which lineages) subject to EDS, then the MEME method is appropriate.
- What about different types of selection?
- Protein sequences can be screened for evidence of directional using the DEPS method, described here, useful when one wants to detect convergent evolution or selective sweeps. For coding sequences, the TOGGLE model, developed by Wayne Delport and colleagues, can detect selection-driven changes that result in amino-acid toggling. A canonical example of this can be found in immune-driven evolution of HIV-1 (escape and reversion).
- Which evolutionary model should I use for my data?
- For each type of data, nucleotide, amino-acid and codon, Datamonkey implements separate model selection procedures. An exhaustive search is performed for all possible (Markov, time-reversible) models of nucleotide evolution. For protein data, a collection of published empirical models are fitted to the alignment and the best one is selected using AICc. Finally, for coding data, a sophisticated genetic-algorithm procedure described in our recent paper is used to examine thousands of potential models and report the best one and various metrics based on the set of credible models - this feature is implemented in the CMS module.
- Did any sites co-evolve?
- A Bayesian graphical model is deduced from reconstructed substitutions at each branch/site combination to infer conditional evolutionary dependancies of sites in the alignments, i.e. whether a site is more or less likely to experience a non-synonymous substitution at a branch when certain other sites do (or do not) experience non-synonymous substitutions at the same branch. The SPIDERMONKEY method was introduced in the evolutionary context in our paper on the evolution of the phenotypically important and highly variable V3 loop of the envelope glycoprotein in HIV-1.
- Has recombination acted upon sequences in an alignment?
- Recombination leaves an imprint on sequence alignments: different segments of the alignment may be described by different phylogenetic trees, called phylogenetic discordance. Datamonkey.org implementes two methods: SBP, suitable for answering the question "Is there evidence of recombination in the alignment?", and GARD, that attempts to find all the recombination breakpoints. Both method are described in this paper. The output of GARD is accepted by most other analyses, and because recombination can mislead phylogenetic analysis that do not account for it, we strongly urge that recombination testing be done on any alignment that is going to be analyzed for positive selection.You can also submit a collection of HIV-1 sequences for recombination screening by a specialized recombination detection algorithm SCUEAL described in this paper.
- What were the ancestral sequences?
- The ASR module implements three different approaches to reconstructing ancestral sequences: joint, marginal and sampled - see this paper for a description and original methodology attribution, from simple or partitioned alignments.
A random effects branch-site model for detecting episodic diversifying selection
http://mbe.oxfordjournals.org/content/early/2011/06/11/molbev.msr125.abstract
订阅:
博文 (Atom)