http://gettinggeneticsdone.blogspot.com/2013/10/useful-linux-oneliners-for-bioinformatics.html
Much of the work that bioinformaticians do is munging and wrangling around massive amounts of text. While there are some "standardized" file formats (FASTQ, SAM, VCF, etc.) and some tools for manipulating them (fastx toolkit, samtools, vcftools, etc.), there are still times where knowing a little bit of Unix/Linux is extremely helpful, namely awk, sed, cut, grep, GNU parallel, and others.
This is by no means an exhaustive catalog, but I've put together a short list of examples using various Unix/Linux utilities for text manipulation, from the very basic (e.g., sum a column) to the very advanced (munge a FASTQ file and print the total number of reads, total number unique reads, percentage of unique reads, most abundant sequence, and its frequency). Most of these examples (with the exception of the SeqTK examples) use built-in utilities installed on nearly every Linux system. These examples are a combination of tactics I used everyday and examples culled from other sources listed at the top of the page.
The list is available as a README in this GitHub repo. This list is a start - I would love suggestions for other things to include. To make a suggestion, leave a comment here, or better - open an issue, or even better still - send me a pull request.
Useful one-liners for bioinformatics: https://github.com/stephenturner/oneliners
Alternatively, download a PDF here.
2013年11月8日星期五
2013年9月22日星期日
Population genomics from pool sequencing
Keywords:
- Pool sequencing;
- High throughput sequencing;
- Neutrality tests;
- Composite likelihood estimators;
- Genetic differentiation
Abstract
Next generation sequencing of pooled samples is an effective approach for studies of variability and differentiation in populations. In this paper we provide a comprehensive set of estimators of the most common statistics in population genetics based on the frequency spectrum, namely the Watterson estimator θW, nucleotide pairwise diversity II, Tajima's D, Fu and Li's D and F, Fay and Wu's H, McDonald-Kreitman and HKA tests and Fst, corrected for sequencing errors and ascertainment bias. In a simulation study, we show that pool and individual θ estimates are highly correlated and discuss how the performance of the statistics vary with read depth and sample size in different evolutionary scenarios. As an application, we reanalyze sequences from Drosophila mauritiana and from an evolution experiment in Drosophila melanogaster. These methods are useful for population genetic projects with limited budget, study of communities of individuals that are hard to isolate, or autopolyploid species.
2013年9月16日星期一
2013年8月14日星期三
tutorials for genomics, phylogenetics, and population genetics
http://evomics.org/learning/all/
This page lists all of the learning activities available on this site.
Workshop Tutorials
Genomics
- Assembly
- AUGUSTUS
- Galaxy
- GBrowse
- Quality Assessment and Quality Control of Sequence Data
- Scripture
- Stacks
Phylogenetics
Population Genetics
Programming
Bioinformatics
2013年7月15日星期一
Java utilities for NGS - Jvarkit
https://github.com/lindenb/jvarkit#vcfgeneontology
http://plindenbaum.blogspot.ca/
Example:
Example:
http://plindenbaum.blogspot.ca/
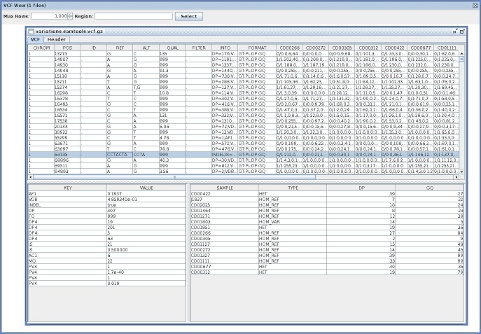
VcfViewGui
VcfViewGui : a Simple java-Swing-based VCF viewer.VCFGeneOntology
vcfgo reads a VCF annotated with VEP or SNPEFF, loads the data from GeneOntology andGOA and adds a new field in the INFO column for the GO terms for each position.Example:
$ java -jar dist/vcfgo.jar I="https://raw.github.com/arq5x/gemini/master/test/tes.snpeff.vcf" |\
grep -v -E '^##' | head -n 3
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT 1094PC0005 1094PC0009 1094PC0012 1094PC0013
chr1 30860 . G C 33.46 . AC=2;AF=0.053;AN=38;BaseQRankSum=2.327;DP=49;Dels=0.00;EFF=DOWNSTREAM(MODIFIER||||85|FAM138A|protein_coding|CODING|ENST00000417324|),DOWNSTREAM(MODIFIER|||||FAM138A|processed_transcript|CODING|ENST00000461467|),DOWNSTREAM(MODIFIER|||||MIR1302-10|miRNA|NON_CODING|ENST00000408384|),INTRON(MODIFIER|||||MIR1302-10|antisense|NON_CODING|ENST00000469289|),INTRON(MODIFIER|||||MIR1302-10|antisense|NON_CODING|ENST00000473358|),UPSTREAM(MODIFIER|||||WASH7P|unprocessed_pseudogene|NON_CODING|ENST00000423562|),UPSTREAM(MODIFIER|||||WASH7P|unprocessed_pseudogene|NON_CODING|ENST00000430492|),UPSTREAM(MODIFIER|||||WASH7P|unprocessed_pseudogene|NON_CODING|ENST00000438504|),UPSTREAM(MODIFIER|||||WASH7P|unprocessed_pseudogene|NON_CODING|ENST00000488147|),UPSTREAM(MODIFIER|||||WASH7P|unprocessed_pseudogene|NON_CODING|ENST00000538476|);FS=3.128;HRun=0;HaplotypeScore=0.6718;InbreedingCoeff=0.1005;MQ=36.55;MQ0=0;MQRankSum=0.217;QD=16.73;ReadPosRankSum=2.017 GT:AD:DP:GQ:PL 0/0:7,0:7:15.04:0,15,177 0/0:2,0:2:3.01:0,3,39 0/0:6,0:6:12.02:0,12,143 0/0:4,0:4:9.03:0,9,119
chr1 69270 . A G 2694.18 . AC=40;AF=1.000;AN=40;DP=83;Dels=0.00;EFF=SYNONYMOUS_CODING(LOW|SILENT|tcA/tcG|S60|305|OR4F5|protein_coding|CODING|ENST00000335137|exon_1_69091_70008);FS=0.000;GOA=OR4F5|GO:0004984&GO:0005886&GO:0004930&GO:0016021;HRun=0;HaplotypeScore=0.0000;InbreedingCoeff=-0.0598;MQ=31.06;MQ0=0;QD=32.86 GT:AD:DP:GQ:PL ./. ./. 1/1:0,3:3:9.03:106,9,0 1/1:0,6:6:18.05:203,18,0
VCFFilterGeneOntology
vcffiltergo reads a VCF annotated with VEP or SNPEFF, loads the data from GeneOntologyand GOA and adds a filter in the FILTER column if a gene at the current genomic location is a descendant of a given GO term.Example:
$ java -jar dist/vcffiltergo.jar I="https://raw.github.com/arq5x/gemini/master/test/test1.snpeff.vcf" \
CHILD_OF=GO:0005886 FILTER=MEMBRANE |\
grep -v "^##" | head -n 3
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT 1094PC0005 1094PC0009 1094PC0012 1094PC0013
chr1 30860 . G C 33.46 PASS AC=2;AF=0.053;AN=38;BaseQRankSum=2.327;DP=49;Dels=0.00;EFF=DOWNSTREAM(MODIFIER||||85|FAM138A|protein_coding|CODING|ENST00000417324|),DOWNSTREAM(MODIFIER|||||FAM138A|processed_transcript|CODING|ENST00000461467|),DOWNSTREAM(MODIFIER|||||MIR1302-10|miRNA|NON_CODING|ENST00000408384|),INTRON(MODIFIER|||||MIR1302-10|antisense|NON_CODING|ENST00000469289|),INTRON(MODIFIER|||||MIR1302-10|antisense|NON_CODING|ENST00000473358|),UPSTREAM(MODIFIER|||||WASH7P|unprocessed_pseudogene|NON_CODING|ENST00000423562|),UPSTREAM(MODIFIER|||||WASH7P|unprocessed_pseudogene|NON_CODING|ENST00000430492|),UPSTREAM(MODIFIER|||||WASH7P|unprocessed_pseudogene|NON_CODING|ENST00000438504|),UPSTREAM(MODIFIER|||||WASH7P|unprocessed_pseudogene|NON_CODING|ENST00000488147|),UPSTREAM(MODIFIER|||||WASH7P|unprocessed_pseudogene|NON_CODING|ENST00000538476|);FS=3.128;HRun=0;HaplotypeScore=0.6718;InbreedingCoeff=0.1005;MQ=36.55;MQ0=0;MQRankSum=0.217;QD=16.73;ReadPosRankSum=2.017 GT:AD:DP:GQ:PL 0/0:7,0:7:15.04:0,15,177 0/0:2,0:2:3.01:0,3,39 0/0:6,0:6:12.02:0,12,143 0/0:4,0:4:9.03:0,9,119
chr1 69270 . A G 2694.18 MEMBRANE AC=40;AF=1.000;AN=40;DP=83;Dels=0.00;EFF=SYNONYMOUS_CODING(LOW|SILENT|tcA/tcG|S60|305|OR4F5|protein_coding|CODING|ENST00000335137|exon_1_69091_70008);FS=0.000;HRun=0;HaplotypeScore=0.0000;InbreedingCoeff=-0.0598;MQ=31.06;MQ0=0;QD=32.86 GT:AD:DP:GQ:PL ./. ./. 1/1:0,3:3:9.03:106,9,0 1/1:0,6:6:18.05:203,18,0
2013年6月27日星期四
Piping with samtools, bwa and bedtools
http://www.biostars.org/p/43677/
In this tutorial I will introduce some concepts related to unix piping. Piping is a very useful feature to avoid creation of intermediate use once files. It is assumed that bedtools, samtools, and bwa are installed.
Lets begin with a typical command to do paired end mapping with bwa: (./ means look in current directory only)
#-t 4 is for using 4 threads/cores
bwa aln -t 4 ./hg19.fasta ./s1_1.fastq > ./s1_1.sai
bwa aln -t 4 ./hg19.fasta ./s1_2.fastq > ./s1_2.sai
bwa sampe ./hg19.fasta ./s1_1.sai ./s1_2.sai ./s1_1.fastq ./s1_2.fastq > s1.sam
samtools view -Shu s1.sam > s1.bam
samtools sort s1.bam s1_sorted
samtools rmdup -s s1_sorted.bam s1_sorted_nodup.bam
bamToBed -i s1_sorted_nodup.bam > s1_sorted_nodup.bed
This workflow above creates many files that are only used once (such as s1.bam) and we can use the unix pipe utility to reduce the number intermediate files created. The pipe function is the character | and what it does is take the output from one command and sets it as input for next command (assuming next command knows how to deal with this input).
For example, when you type in
head myfile.txt the command 'head' will read first 10 lines of myfile.txt and output it (by default) to the display (stdout) so you will see the first 10 lines printed on your screen. However, if you were to do something like this:head myfile.txt | wc -l
you will instead get
10 printed out on your screen. What the pipe command did was take the output of head command and used it as input for the wordcount (wc) command. wc -l command will count the number of lines passed in (which is 10 since head by default prints the first 10 lines). An equivalent command would behead myfile.txt > myfile_top10_lines.txt
wc -l myfile_top10_lines.txt
By using the pipe command we are able to eliminate the intermediate file that was generated (myfile_top10_lines.txt).
Below is an example of how pipe command can be used in samtools
bwa aln -t 4 ./hg19.fasta ./s1_1.fastq > ./s1_1.sai
bwa aln -t 4 ./hg19.fasta ./s1_2.fastq > ./s1_2.sai
bwa sampe ./hg19.fasta ./s1_1.sai ./s1_2.sai ./s1_1.fastq ./s1_2.fastq | \
samtools view -Shu - | \
samtools sort - - | \
samtools rmdup -s - - | \
tee s1_sorted_nodup.bam | \
bamToBed > s1_sorted_nodup.bed
A couple of things I wanted to point out here:
- The \ symbol after the pipe symbol tells the terminal that I want to keep writing command on the next line, that way you can have multiline commands instead of super-long command on one line.
- The tee command takes the information being piped and creates a file while passing the data along the pipe, this command is useful if you want to save an intermediate file but still need to do additional processing. (Think of a T shaped drain pipe)
- The - symbol in samtools it to tell the samtools program to take the input from pipe
- It is not recommended you run this command all at once like I showed above if you have a giant bam file because
sortwill take forever. Running things in a pipe can be more space efficient since you are not storing intermediate files but can create issues/bottlenecks elsewhere. If problems arise, run the pieces without piping to troubleshoot. - The two bwa aln commands can be run in parallel (for example: on another machine) before the sampe command
Other common uses of pipes that I have not shown above it to pipe output into snp calling, you can find some examples here: (NOTE: pileupcommand is depreciated, the new way to call SNPs using samtools using mpileup command)
samtools merge - *.bam | tee merged.bam | samtools rmdup - - | tee rmdup.bam | samtools mpileup - uf ./hg19.fasta - | bcftools view -bvcg - | gzip > var.raw.bcf.gz
Compressing files with gzip can save a lot of space, but makes these files non-human readable (if you use the
head or less command it will give you gibberish). To view these files you can use zless and zcat command.zless var.raw.bcf.gz #similar to gzip -cd var.raw.bcf.gz | less
zcat var.raw.bcf.gz | head
In case you are wondering if there is a way to run bwa sampe without storing intermediate sai files the command below will do this:
bwa sampe ./hg19.fasta <(bwa aln -t 4 ./hg19.fasta ./s1_1.fastq) <(bwa aln -t 4 ./hg19.fasta ./s1_2.fastq) ./s1_1.fastq ./s1_2.fastq | samtools view -Shb /dev/stdin > s1.bam
Notice here that I used the -b command instead of -u command in samtools view. -u is useful for piping since it spits out an uncompressed bam so you don't have to waste CPU cycles compressing/decompressing at every step, however, if you have a file you want to store, it would be better to compress it with -b. Additionally, I used /dev/stdin instead of using the - symbol, they both mean the same thing (afaik) and I wanted to point it out that /dev/stdin might be a more explicit way of showing things. (ie samtools view -Shu /dev/stdin )
2013年5月8日星期三
Sequence logos
1. http://weblogo.berkeley.edu/logo.cgi
2. generate logo from alignment using R
http://davetang.org/muse/2013/01/30/sequence-logos-with-r/
2. generate logo from alignment using R
http://davetang.org/muse/2013/01/30/sequence-logos-with-r/
2013年5月1日星期三
2013年4月24日星期三
DNAPlotter: circular and linear interactive genome visualization
http://bioinformatics.oxfordjournals.org/content/25/1/119.full
Circular and linear DNA diagrams provide a powerful tool for illustrating the features of a genome in their full genomic context. CGView (Stothard and Wishart,2005) is a circular genome viewer that produces PNG, JPEG or scalable vector graphics (SVG) format. It is primarily designed to be part of a pipeline and not an interactive and editable viewer. GenomeDiagram (Pritchard et al., 2006) is a Python module, which can also be used to generate diagrams. GenomePlot (Gibson and Smith, 2003), GenoMap (Sato and Ehira, 2003) and Microbial Genome Viewer (MGV; Kerkhoven et al., 2004), are GUI based but do not provide the ability to directly read in files in the common sequence formats or a filtering tool to define tracks.
Circos (http://mkweb.bcgsc.ca/circos/) is web-based genome comparison visualizer that is configured from flat files. It can be configured for comparisons within a genome or between multiple genomes. There are also commercial packages available that draw circular and linear plots.
Below we present DNAPlotter, a collaborative project combining graphics from the Jemboss (http://emboss.sf.net/Jemboss/) DNA viewer and Artemis (http://www.sanger.ac.uk/Software/Artemis/) libraries to read files and filter features, which has been designed to be a graphical means of customizing circular and linear diagrams.
2013年4月23日星期二
Crossing the species barrier: genomic hotspots of introgression between two highly divergent Ciona intestinalis species
http://mbe.oxfordjournals.org/content/early/2013/04/05/molbev.mst066.full.pdf+html
ABC running with sliding-windows
Inferring a realistic demographic model from genetic data is an important challenge to gain insights into the historical events during the speciation process and to detect molecular signatures of selection along genomes. Recent advances in divergence population genetics have reported that speciation in face of gene flow occurred more frequently than theoretically expected, but the approaches used did not account for genome wide heterogeneity (GWH) in introgression rates. Here we investigate the impact of GWH on the inference of divergence with gene flow between two cryptic species of the marine modelCiona intestinalis by analysing polymorphism and divergence patterns in 852 protein-coding sequence loci.
These morphologically similar entities are highly diverged molecular-wise, but evidence of hybridisation has been reported in both laboratory and field studies. We compare various speciation models and test for GWH under the ABC framework. Our results demonstrate the presence of significant extents of gene flow resulting from a recent secondary contact after >3 My of divergence in isolation. The inferred rates of introgression are relatively low, highly variable across loci and mostly unidirectional, which is consistent with the idea that numerous genetic incompatibilities have accumulated over time throughout the genomes of these highly-diverged species. A genomic map of the level of gene flow identified two hotspots of introgression, i.e. large genome regions of unidirectional introgression. This study clarifies the history and degree of isolation of two cryptic and partially sympatric model species, and provides a methodological framework to investigate GWH at various stages of speciation process.
CodonPhyML: Fast Maximum Likelihood Phylogeny Estimation under Codon Substitution Models
http://mbe.oxfordjournals.org/content/early/2013/04/02/molbev.mst034.abstract
Markov models of codon substitution naturally incorporate the structure of the genetic code and the selection intensity at the protein level, providing a more realistic representation of protein-coding sequences compared with nucleotide or amino acid models. Thus, for protein-coding genes, phylogenetic inference is expected to be more accurate under codon models. So far, phylogeny reconstruction under codon models has been elusive due to computational difficulties of dealing with high dimension matrices. Here, we present a fast maximum likelihood (ML) package for phylogenetic inference, CodonPhyML offering hundreds of different codon models, the largest variety to date, for phylogeny inference by ML. CodonPhyML is tested on simulated and real data and is shown to offer excellent speed and convergence properties. In addition, CodonPhyML includes most recent fast methods for estimating phylogenetic branch supports and provides an integral framework for models selection, including amino acid and DNA models.
Markov models of codon substitution naturally incorporate the structure of the genetic code and the selection intensity at the protein level, providing a more realistic representation of protein-coding sequences compared with nucleotide or amino acid models. Thus, for protein-coding genes, phylogenetic inference is expected to be more accurate under codon models. So far, phylogeny reconstruction under codon models has been elusive due to computational difficulties of dealing with high dimension matrices. Here, we present a fast maximum likelihood (ML) package for phylogenetic inference, CodonPhyML offering hundreds of different codon models, the largest variety to date, for phylogeny inference by ML. CodonPhyML is tested on simulated and real data and is shown to offer excellent speed and convergence properties. In addition, CodonPhyML includes most recent fast methods for estimating phylogenetic branch supports and provides an integral framework for models selection, including amino acid and DNA models.
2013年4月21日星期日
a Little Book of R for Bioinformatics
http://a-little-book-of-r-for-bioinformatics.readthedocs.org/en/latest/#
Chapters in this Book
- How to install R and a Brief Introduction to R
- Neglected Tropical diseases
- DNA Sequence Statistics (1)
- DNA Sequence Statistics (2)
- Sequence Databases
- REVISION EXERCISES 1
- Pairwise Sequence Alignment
- Multiple Alignment and Phylogenetic trees
- REVISION EXERCISES 2
- Computational Gene-finding
- Comparative Genomics
- Hidden Markov Models
- Answers to the End-of-chapter Exercises
- Answers to Revision Exercises
- Protein-Protein Interaction Graphs
2013年4月17日星期三
A wide extent of inter-strain diversity in virulent and vaccine strains of alpha-herpesviruses
Good example on de novo genome assembly, annotation, comparsion
http://genomics-pubs.princeton.edu/prv/scripts.shtml
http://genomics-pubs.princeton.edu/prv/scripts.shtml
Overview & Table of Contents
Part I - Pre-Assembly Processing
These steps take raw Illumina® sequence data and prepare it for de novo assembly, using a series of quality-controls and filters.
Part II - De novo Assembly & Genome Finishing
This phase of genome production takes high-quality short sequence reads, and by homology-based overlaps, joins them into longer and longer pieces.
Part III - Quality Assessment of New Genomes
These steps assess the quality of the assembly and address any inconsistencies. These scripts function in conjunction with wet-bench analyses, such as restriction fragment length polymorphism (RFLP) analysis and directed PCR validations.
Part IV - Annotation & Genome Comparison
This phase involves annotation, curation, and comparison of multiple genome sequences to reveal interesting biological features.
Addendum - Relevant Websites and Lists of Scripts Used
All scripts used are freely available online (web links provided) or are available here (see Downloads link on left) for download and free use.
2013年4月7日星期日
Eckertlab - on evolutionary genomics of trees
http://eckertlab.blogspot.ca/
the Eckert Lab located in the Department of Biology at Virginia Commonwealth University.
A plant evolutionary genomics lab.
Research topics range from the dissection of adaptive plant phenotypes into their genetic components to inferences of genome-wide patterns of polymorphism, divergence and natural selection.
http://evolfri.blogspot.ca/
the Eckert Lab located in the Department of Biology at Virginia Commonwealth University.
A plant evolutionary genomics lab.
Research topics range from the dissection of adaptive plant phenotypes into their genetic components to inferences of genome-wide patterns of polymorphism, divergence and natural selection.
http://evolfri.blogspot.ca/
2013年4月6日星期六
good GUI tool for sequences analysis
1. Unipro UGENE
http://ugene.unipro.ru/
http://bioinformatics.oxfordjournals.org/content/28/8/1166
http://ugene.unipro.ru/
http://bioinformatics.oxfordjournals.org/content/28/8/1166
- CAP3
- Bowtie-0.12.7
- BWA-0.5.9
- blast-2.2.25
- blast-2.2.25+
- clustalw-2.1
- mafft-6.847
- T-Coffee-8.99
- Mr.Bayes-3.2.0
Summary: Unipro UGENE is a multiplatform open-source software with the main goal of assisting molecular biologists without much expertise in bioinformatics to manage, analyze and visualize their data. UGENE integrates widely used bioinformatics tools within a common user interface. The toolkit supports multiple biological data formats and allows the retrieval of data from remote data sources. It provides visualization modules for biological objects such as annotated genome sequences, Next Generation Sequencing (NGS) assembly data, multiple sequence alignments, phylogenetic trees and 3D structures. Most of the integrated algorithms are tuned for maximum performance by the usage of multithreading and special processor instructions. UGENE includes a visual environment for creating reusable workflows that can be launched on local resources or in a High Performance Computing (HPC) environment. UGENE is written in C++ using the Qt framework. The built-in plugin system and structured UGENE API make it possible to extend the toolkit with new functionality.
Availability and implementation: UGENE binaries are freely available for MS Windows, Linux and Mac OS X at http://ugene.unipro.ru/download.html. UGENE code is licensed under the GPLv2; the information about the code licensing and copyright of integrated tools can be found in the LICENSE.3rd_party file provided with the source bundle.
2013年4月2日星期二
modify sequence name in a fasta file
cat genes.fasta | awk '{if (/^>/) {print $1} else {print $0}}' > genes2.fasta
To shorten the sequence names in fasta file.
change
>sequense1 human person1
ATGC
to
>sequense1
ATGC
To shorten the sequence names in fasta file.
change
>sequense1 human person1
ATGC
to
>sequense1
ATGC
Some unix/perl oneliners for Bioinformatics
http://genomics-array.blogspot.ca/2010/11/some-unixperl-oneliners-for.html
1 File format conversion/line counting/counting number of files etc.
1. $ wc –l : count number of lines in a file.
2. $ ls | wc –l : count number of files in a directory.
3. $ tac : print the file in reverse order e.g; last line first, first line last.
4. $ rev : reverse the file in lines.
5. $ sed 's/.$//' or sed 's/^M$//' or sed 's/\x0D$//' : converts a dos file into unix mode.
6. $sed "s/$/`echo -e \\\r`/" or sed 's/$/\r/' or sed "s/$//": converts a unix newline into a DOS newline.
7. $ awk '1; { print "" }' : Double space a file.
8. $ awk '{ total = total + NF }; END { print total+0 }' : prints the number of words in a file.
9. $sed '/^$/d' or [grep ‘.’] : Delete all blank lines in a file.
10. $sed '/./,$!d' : Delete all blank lines in the beginning of the file.
11. $sed -e :a -e '/^\n*$/{$d;N;ba' -e '}': Delete all blank lines at the end of the file.
12. $sed -e :a -e 's/<[^>]*>//g;/
13. $sed 's/^[ \t]*//' : deleting all leading white space tabs in a file.
14. $ sed 's/[ \t]*$//' : Delete all trailing white space and tab in a file.
15. $ sed 's/^[ \t]*//;s/[ \t]*$//' : Delete both leading and trailing white space and tab in a file.
2.2 Working with Patterns/numbers in a sequence file
16. $awk '/Pattern/ { n++ }; END { print n+0 }' : print the total number of lines containing the word pattern.
17. $sed 10q : print first 10 lines.
18. $sed -n '/regexp/p' : Print the line that matches the pattern.
19. $sed '/regexp/d' : Deletes the lines that matches the regexp.
20. $sed -n '/regexp/!p' : Print the lines that does not match the pattern.
21. $sed '/regexp/!d' : Deletes the lines that does NOT match the regular expression.
22. $sed -n '/^.\{65\}/p' : print lines that are longer than 65 characters.
23. $sed -n '/^.\{65\}/!p' : print lines that are lesser than 65 characters.
24. $sed -n '/regexp/{g;1!p;};h' : print one line before the pattern match.
25. $sed -n '/regexp/{n;p;}' : print one line after the pattern match.
26. $sed -n '/^.\{65\}/ {g;1!p;};h' < sojae_seq > tmp : print the names of the sequences that are larger than 65 nucleotide long.
27. $sed -n '/regexp/,$p' : Print regular expression to the end of file.
28. $sed -n '8,12p' : print line 8 to 12(inclusive)
29. $sed -n '52p' : print only line number 52.
30. $seq ‘/pattern1/,/pattern2/d’ < inputfile > outfile : will delete all the lines between pattern1 and pattern2.
31. $sed ‘/20,30/d’ < inputfile > outfile : will delete all lines between 20 and 30. OR sed ‘/20,30/d’ < input > output will delete lines between 20 and 30.
32. awk '/baz/ { gsub(/foo/, "bar") }; { print }' : Substitute foo with bar in lines that contains ‘baz’.
33. awk '!/baz/ { gsub(/foo/, "bar") }; { print }' : Substitute foo with bar in lines that does not contain ‘baz’.
34. grep –i –B 1 ‘pattern’ filename > out : Will print the name of the sequence and the sequence having the pattern in a case insensitive way(make sure the sequence name and the sequence each occupy a single line).
35. grep –i –A 1 ‘seqname’ filename > out : will print the sequence name as well as the sequence into file ‘out’.
2.3 Inserting Data into a file:
36. gawk --re-interval 'BEGIN{ while(a++<49 1="" amp="" filename="" nbsp="" s="" sub="" x="">> fileout : will insert 49 ‘X’ in the sixth position of every line.
37. gawk --re-interval 'BEGIN{ s="YourName" }; { sub(/^.{6}/,"&" s) }; 1' : Insert your name at the 6 th position in every line.
3. Working with Data Files[Tab delimited files]:
3.1 Error Checking and data handling:
38. awk '{ print NF ":" $0 } ' : print the number of fields of each line followed by the line.
39. awk '{ print $NF }' : print the last field of each line.
40. awk 'NF > n' : print every line with more than ‘n’ fields.
41. awk '$NF > n' : print every line where the last field is greater than n.
42. awk '{ print $2, $1 }': prints just first 2 fields of a data file in reverse order.
43. awk '{ temp = $1; $1 = $2; $2 = temp; print }' : prints all the fields in the correct order except the first 2 fields.
44. awk '{ for (i=NF; i>0; i--) printf("%s ", $i); printf ("\n") }' : prints all the fields in reverse order.
45. awk '{ $2 = ""; print }' : deletes the 2nd field in each line.
46. awk '$5 == "abc123"' : print each line where the 5th field is equal to ‘abc123’.
47. awk '$5 != "abc123"' : print each line where 5th field is NOT equal to abc123.
48. awk '$7 ~ /^[a-f]/' : Print each line whose 7th field matches the regular expression.
49. awk '$7 !~ /^[a-f]/' : print each line whose 7th field does NOT match the regular expression.
50. cut –f n1,n2,n3..> output file : will cut n1,n2,n3 columns(fields) from input file and print the output in output file. If delimiter is other than TAB then give additional argument such as cut –d ‘,’ –f n1,n2.. inputfile > out
51. sort –n –k 2,2 –k 4,4 file > fileout : Will conduct a numerical sort of column 2, and then column 4. If –n is not specified, then, sort will do a lexicographical sort(of the ascii value).
4. Miscellaneous:
52. uniq –u inputfile > out : will print only the uniq lines present in the sorted input file.
53. uniq –d inputfile > out : will print only the lines that are in doubles from the sorted input file.
54. cat file1 file2 file3 … fileN > outfile : Will concatenate files back to back in outfile.
55. paste file1 file2 > outfile : will merge two files horizontally. This function is good for merging with same number of rows but different column width.
56. !:p : will print the previous command run with the ‘pattern’ in it.
57. !! : repeat the last command entered at the shell.
58. ~ : Go back to home directory
59. echo {a,t,g,c}{a,t,g,c}{a,t,g,c}{a,t,g,c} : will generate all tetramers using ‘atgc’. If you want pentamers/hexamers etc. then just increase the number of bracketed entities.NOTE: This is not a efficient sequence shuffler. If you wish to generate longer sequences then use other means.
60. kill -HUP ` ps -aef | grep -i firefox | sort -k 2 -r | sed 1d | awk ' { print $2 } ' ` : Kills a hanging firefox process.
61. csplit -n 7 input.fasta '/>/' '{*}' : will split the file ‘input.fasta’ wherever it encounters delimiter ‘>’. The file names will appear as 7 digit long strings.
62. find . -name data.txt –print: finds and prints the path for file data.txt.
Sample Script to make set operations on sequence files:
63. grep ‘>’ filenameA > list1 # Will list just the sequence names in a file names.
grep ‘>’ filenameB > list2 # Will list names for file 2
cat list1 list2 > tmp # concatenates list1 and list2 into tmp
sort tmp > tmp1 # File sorted
uniq –u tmp1 > uniq # AUB – A ∩ B (OR (A-B) U (B-A))
uniq –d tmp1 > double # Is the intersection (A ∩ B)
cat uniq double > Union # AUB
cat list1 double > tmp
sort tmp | uniq –u > list1uniq # A - B
cat list2 double > tmp
sort tmp | uniq –u > list2uniq # B - A
PERL ONELINERS:
1. perl -pe '$\="\n"' : double space a file
2. perl -pe '$_ .= "\n" unless /^$/' : double space a file except blank lines
3. perl -pe '$_.="\n"x7' : 7 space in a line.
4. perl -ne 'print unless /^$/' : remove all blank lines
5. perl -lne 'print if length($_) < 20' : print all lines with length less than 20.
6. perl -00 -pe '' : If there are multiple spaces, delete all leaving one(make the file a single spaced file).
7. perl -00 -pe '$_.="\n"x4' : Expand single blank lines into 4 consecutive blank lines
8. perl -pe '$_ = "$. $_"': Number all lines in a file
9. perl -pe '$_ = ++$a." $_" if /./' : Number only non-empty lines in a file
10. perl -ne 'print ++$a." $_" if /./' : Number and print only non-empty lines in a file
11. perl -pe '$_ = ++$a." $_" if /regex/' ; Number only lines that match a pattern
12. perl -ne 'print ++$a." $_" if /regex/' : Number and print only lines that match a pattern
13. perl -ne 'printf "%-5d %s", $., $_ if /regex/' : Left align lines with 5 white spaces if matches a pattern (perl -ne 'printf "%-5d %s", $., $_' : for all the lines)
14. perl -le 'print scalar(grep{/./}<>)' : prints the total number of non-empty lines in a file
15. perl -lne '$a++ if /regex/; END {print $a+0}' : print the total number of lines that matches the pattern
16. perl -alne 'print scalar @F' : print the total number fields(words) in each line.
17. perl -alne '$t += @F; END { print $t}' : Find total number of words in the file
18. perl -alne 'map { /regex/ && $t++ } @F; END { print $t }' : find total number of fields that match the pattern
19. perl -lne '/regex/ && $t++; END { print $t }' : Find total number of lines that match a pattern
20. perl -le '$n = 20; $m = 35; ($m,$n) = ($n,$m%$n) while $n; print $m' : will calculate the GCD of two numbers.
21. perl -le '$a = $n = 20; $b = $m = 35; ($m,$n) = ($n,$m%$n) while $n; print $a*$b/$m' : will calculate lcd of 20 and 35.
22. perl -le '$n=10; $min=5; $max=15; $, = " "; print map { int(rand($max-$min))+$min } 1..$n' : Generates 10 random numbers between 5 and 15.
23. perl -le 'print map { ("a".."z",”0”..”9”)[rand 36] } 1..8': Generates a 8 character password from a to z and number 0 – 9.
24. perl -le 'print map { ("a",”t”,”g”,”c”)[rand 4] } 1..20': Generates a 20 nucleotide long random residue.
25. perl -le 'print "a"x50': generate a string of ‘x’ 50 character long
26. perl -le 'print join ", ", map { ord } split //, "hello world"': Will print the ascii value of the string hello world.
27. perl -le '@ascii = (99, 111, 100, 105, 110, 103); print pack("C*", @ascii)': converts ascii values into character strings.
28. perl -le '@odd = grep {$_ % 2 == 1} 1..100; print "@odd"': Generates an array of odd numbers.
29. perl -le '@even = grep {$_ % 2 == 0} 1..100; print "@even"': Generate an array of even numbers
30. perl -lpe 'y/A-Za-z/N-ZA-Mn-za-m/' file: Convert the entire file into 13 characters offset(ROT13)
31. perl -nle 'print uc' : Convert all text to uppercase:
32. perl -nle 'print lc' : Convert text to lowercase:
33. perl -nle 'print ucfirst lc' : Convert only first letter of first word to uppercas
34. perl -ple 'y/A-Za-z/a-zA-Z/' : Convert upper case to lower case and vice versa
35. perl -ple 's/(\w+)/\u$1/g' : Camel Casing
36. perl -pe 's|\n|\r\n|' : Convert unix new lines into DOS new lines:
37. perl -pe 's|\r\n|\n|' : Convert DOS newlines into unix new line
38. perl -pe 's|\n|\r|' : Convert unix newlines into MAC newlines:
39. perl -pe '/regexp/ && s/foo/bar/' : Substitute a foo with a bar in a line with a regexp.
Some other Perl Tricks
Want to display some progress bars while perl does your job:
For this perl provides a nice utility called "pipe opens" ('perldoc -f open' will provide more info)
1 File format conversion/line counting/counting number of files etc.
1. $ wc –l
2. $ ls | wc –l : count number of files in a directory.
3. $ tac
4. $ rev
5. $ sed 's/.$//' or sed 's/^M$//' or sed 's/\x0D$//' : converts a dos file into unix mode.
6. $sed "s/$/`echo -e \\\r`/" or sed 's/$/\r/' or sed "s/$//": converts a unix newline into a DOS newline.
7. $ awk '1; { print "" }' : Double space a file.
8. $ awk '{ total = total + NF }; END { print total+0 }' : prints the number of words in a file.
9. $sed '/^$/d' or [grep ‘.’] : Delete all blank lines in a file.
10. $sed '/./,$!d' : Delete all blank lines in the beginning of the file.
11. $sed -e :a -e '/^\n*$/{$d;N;ba' -e '}': Delete all blank lines at the end of the file.
12. $sed -e :a -e 's/<[^>]*>//g;/
13. $sed 's/^[ \t]*//' : deleting all leading white space tabs in a file.
14. $ sed 's/[ \t]*$//' : Delete all trailing white space and tab in a file.
15. $ sed 's/^[ \t]*//;s/[ \t]*$//' : Delete both leading and trailing white space and tab in a file.
2.2 Working with Patterns/numbers in a sequence file
16. $awk '/Pattern/ { n++ }; END { print n+0 }' : print the total number of lines containing the word pattern.
17. $sed 10q : print first 10 lines.
18. $sed -n '/regexp/p' : Print the line that matches the pattern.
19. $sed '/regexp/d' : Deletes the lines that matches the regexp.
20. $sed -n '/regexp/!p' : Print the lines that does not match the pattern.
21. $sed '/regexp/!d' : Deletes the lines that does NOT match the regular expression.
22. $sed -n '/^.\{65\}/p' : print lines that are longer than 65 characters.
23. $sed -n '/^.\{65\}/!p' : print lines that are lesser than 65 characters.
24. $sed -n '/regexp/{g;1!p;};h' : print one line before the pattern match.
25. $sed -n '/regexp/{n;p;}' : print one line after the pattern match.
26. $sed -n '/^.\{65\}/ {g;1!p;};h' < sojae_seq > tmp : print the names of the sequences that are larger than 65 nucleotide long.
27. $sed -n '/regexp/,$p' : Print regular expression to the end of file.
28. $sed -n '8,12p' : print line 8 to 12(inclusive)
29. $sed -n '52p' : print only line number 52.
30. $seq ‘/pattern1/,/pattern2/d’ < inputfile > outfile : will delete all the lines between pattern1 and pattern2.
31. $sed ‘/20,30/d’ < inputfile > outfile : will delete all lines between 20 and 30. OR sed ‘/20,30/d’ < input > output will delete lines between 20 and 30.
32. awk '/baz/ { gsub(/foo/, "bar") }; { print }' : Substitute foo with bar in lines that contains ‘baz’.
33. awk '!/baz/ { gsub(/foo/, "bar") }; { print }' : Substitute foo with bar in lines that does not contain ‘baz’.
34. grep –i –B 1 ‘pattern’ filename > out : Will print the name of the sequence and the sequence having the pattern in a case insensitive way(make sure the sequence name and the sequence each occupy a single line).
35. grep –i –A 1 ‘seqname’ filename > out : will print the sequence name as well as the sequence into file ‘out’.
2.3 Inserting Data into a file:
36. gawk --re-interval 'BEGIN{ while(a++<49 1="" amp="" filename="" nbsp="" s="" sub="" x="">> fileout : will insert 49 ‘X’ in the sixth position of every line.
37. gawk --re-interval 'BEGIN{ s="YourName" }; { sub(/^.{6}/,"&" s) }; 1' : Insert your name at the 6 th position in every line.
3. Working with Data Files[Tab delimited files]:
3.1 Error Checking and data handling:
38. awk '{ print NF ":" $0 } ' : print the number of fields of each line followed by the line.
39. awk '{ print $NF }' : print the last field of each line.
40. awk 'NF > n' : print every line with more than ‘n’ fields.
41. awk '$NF > n' : print every line where the last field is greater than n.
42. awk '{ print $2, $1 }'
43. awk '{ temp = $1; $1 = $2; $2 = temp; print }' : prints all the fields in the correct order except the first 2 fields.
44. awk '{ for (i=NF; i>0; i--) printf("%s ", $i); printf ("\n") }' : prints all the fields in reverse order.
45. awk '{ $2 = ""; print }' : deletes the 2nd field in each line.
46. awk '$5 == "abc123"' : print each line where the 5th field is equal to ‘abc123’.
47. awk '$5 != "abc123"' : print each line where 5th field is NOT equal to abc123.
48. awk '$7 ~ /^[a-f]/' : Print each line whose 7th field matches the regular expression.
49. awk '$7 !~ /^[a-f]/' : print each line whose 7th field does NOT match the regular expression.
50. cut –f n1,n2,n3..
51. sort –n –k 2,2 –k 4,4 file > fileout : Will conduct a numerical sort of column 2, and then column 4. If –n is not specified, then, sort will do a lexicographical sort(of the ascii value).
4. Miscellaneous:
52. uniq –u inputfile > out : will print only the uniq lines present in the sorted input file.
53. uniq –d inputfile > out : will print only the lines that are in doubles from the sorted input file.
54. cat file1 file2 file3 … fileN > outfile : Will concatenate files back to back in outfile.
55. paste file1 file2 > outfile : will merge two files horizontally. This function is good for merging with same number of rows but different column width.
56. !
57. !! : repeat the last command entered at the shell.
58. ~ : Go back to home directory
59. echo {a,t,g,c}{a,t,g,c}{a,t,g,c}{a,t,g,c} : will generate all tetramers using ‘atgc’. If you want pentamers/hexamers etc. then just increase the number of bracketed entities.NOTE: This is not a efficient sequence shuffler. If you wish to generate longer sequences then use other means.
60. kill -HUP ` ps -aef | grep -i firefox | sort -k 2 -r | sed 1d | awk ' { print $2 } ' ` : Kills a hanging firefox process.
61. csplit -n 7 input.fasta '/>/' '{*}' : will split the file ‘input.fasta’ wherever it encounters delimiter ‘>’. The file names will appear as 7 digit long strings.
62. find . -name data.txt –print: finds and prints the path for file data.txt.
Sample Script to make set operations on sequence files:
63. grep ‘>’ filenameA > list1 # Will list just the sequence names in a file names.
grep ‘>’ filenameB > list2 # Will list names for file 2
cat list1 list2 > tmp # concatenates list1 and list2 into tmp
sort tmp > tmp1 # File sorted
uniq –u tmp1 > uniq # AUB – A ∩ B (OR (A-B) U (B-A))
uniq –d tmp1 > double # Is the intersection (A ∩ B)
cat uniq double > Union # AUB
cat list1 double > tmp
sort tmp | uniq –u > list1uniq # A - B
cat list2 double > tmp
sort tmp | uniq –u > list2uniq # B - A
PERL ONELINERS:
1. perl -pe '$\="\n"'
2. perl -pe '$_ .= "\n" unless /^$/' : double space a file except blank lines
3. perl -pe '$_.="\n"x7' : 7 space in a line.
4. perl -ne 'print unless /^$/' : remove all blank lines
5. perl -lne 'print if length($_) < 20' : print all lines with length less than 20.
6. perl -00 -pe '' : If there are multiple spaces, delete all leaving one(make the file a single spaced file).
7. perl -00 -pe '$_.="\n"x4' : Expand single blank lines into 4 consecutive blank lines
8. perl -pe '$_ = "$. $_"': Number all lines in a file
9. perl -pe '$_ = ++$a." $_" if /./' : Number only non-empty lines in a file
10. perl -ne 'print ++$a." $_" if /./' : Number and print only non-empty lines in a file
11. perl -pe '$_ = ++$a." $_" if /regex/' ; Number only lines that match a pattern
12. perl -ne 'print ++$a." $_" if /regex/' : Number and print only lines that match a pattern
13. perl -ne 'printf "%-5d %s", $., $_ if /regex/' : Left align lines with 5 white spaces if matches a pattern (perl -ne 'printf "%-5d %s", $., $_' : for all the lines)
14. perl -le 'print scalar(grep{/./}<>)' : prints the total number of non-empty lines in a file
15. perl -lne '$a++ if /regex/; END {print $a+0}' : print the total number of lines that matches the pattern
16. perl -alne 'print scalar @F' : print the total number fields(words) in each line.
17. perl -alne '$t += @F; END { print $t}' : Find total number of words in the file
18. perl -alne 'map { /regex/ && $t++ } @F; END { print $t }' : find total number of fields that match the pattern
19. perl -lne '/regex/ && $t++; END { print $t }' : Find total number of lines that match a pattern
20. perl -le '$n = 20; $m = 35; ($m,$n) = ($n,$m%$n) while $n; print $m' : will calculate the GCD of two numbers.
21. perl -le '$a = $n = 20; $b = $m = 35; ($m,$n) = ($n,$m%$n) while $n; print $a*$b/$m' : will calculate lcd of 20 and 35.
22. perl -le '$n=10; $min=5; $max=15; $, = " "; print map { int(rand($max-$min))+$min } 1..$n' : Generates 10 random numbers between 5 and 15.
23. perl -le 'print map { ("a".."z",”0”..”9”)[rand 36] } 1..8': Generates a 8 character password from a to z and number 0 – 9.
24. perl -le 'print map { ("a",”t”,”g”,”c”)[rand 4] } 1..20': Generates a 20 nucleotide long random residue.
25. perl -le 'print "a"x50': generate a string of ‘x’ 50 character long
26. perl -le 'print join ", ", map { ord } split //, "hello world"': Will print the ascii value of the string hello world.
27. perl -le '@ascii = (99, 111, 100, 105, 110, 103); print pack("C*", @ascii)': converts ascii values into character strings.
28. perl -le '@odd = grep {$_ % 2 == 1} 1..100; print "@odd"': Generates an array of odd numbers.
29. perl -le '@even = grep {$_ % 2 == 0} 1..100; print "@even"': Generate an array of even numbers
30. perl -lpe 'y/A-Za-z/N-ZA-Mn-za-m/' file: Convert the entire file into 13 characters offset(ROT13)
31. perl -nle 'print uc' : Convert all text to uppercase:
32. perl -nle 'print lc' : Convert text to lowercase:
33. perl -nle 'print ucfirst lc' : Convert only first letter of first word to uppercas
34. perl -ple 'y/A-Za-z/a-zA-Z/' : Convert upper case to lower case and vice versa
35. perl -ple 's/(\w+)/\u$1/g' : Camel Casing
36. perl -pe 's|\n|\r\n|' : Convert unix new lines into DOS new lines:
37. perl -pe 's|\r\n|\n|' : Convert DOS newlines into unix new line
38. perl -pe 's|\n|\r|' : Convert unix newlines into MAC newlines:
39. perl -pe '/regexp/ && s/foo/bar/' : Substitute a foo with a bar in a line with a regexp.
Some other Perl Tricks
Want to display some progress bars while perl does your job:
For this perl provides a nice utility called "pipe opens" ('perldoc -f open' will provide more info)
open(my $file, '-|', 'command','option', 'option', ...) or die "Could not run tar ... - $!";
while (<$file>) {
print "-";
}
print "\n";
close($file); Will print - on the screen till the process is completed
订阅:
评论 (Atom)