I am the head of the Computational and Mathematical Biology group in Grenoble. My research interests are in statistical population genetics and evolutionary genomics. I develop computational and statistical methods for the inference of population structure, demography and local adaptation from genomic data. My approaches are mainly based on MCMC, machine learning and approximate Bayesian approaches, including geographic and environmental data. My goal is to provide improved statistical estimation procedures under equilibrium and non-equilibrium processes in population genetics. I also study applications in coalescent theory, the mathematical properties of genealogies and the shape of phylogenetic trees.
My group distributes the computer programs TESS and POPS which compute individual cluster membership probabilities and admixture coefficients using multilocus genotypes and geographical or environmental variables.
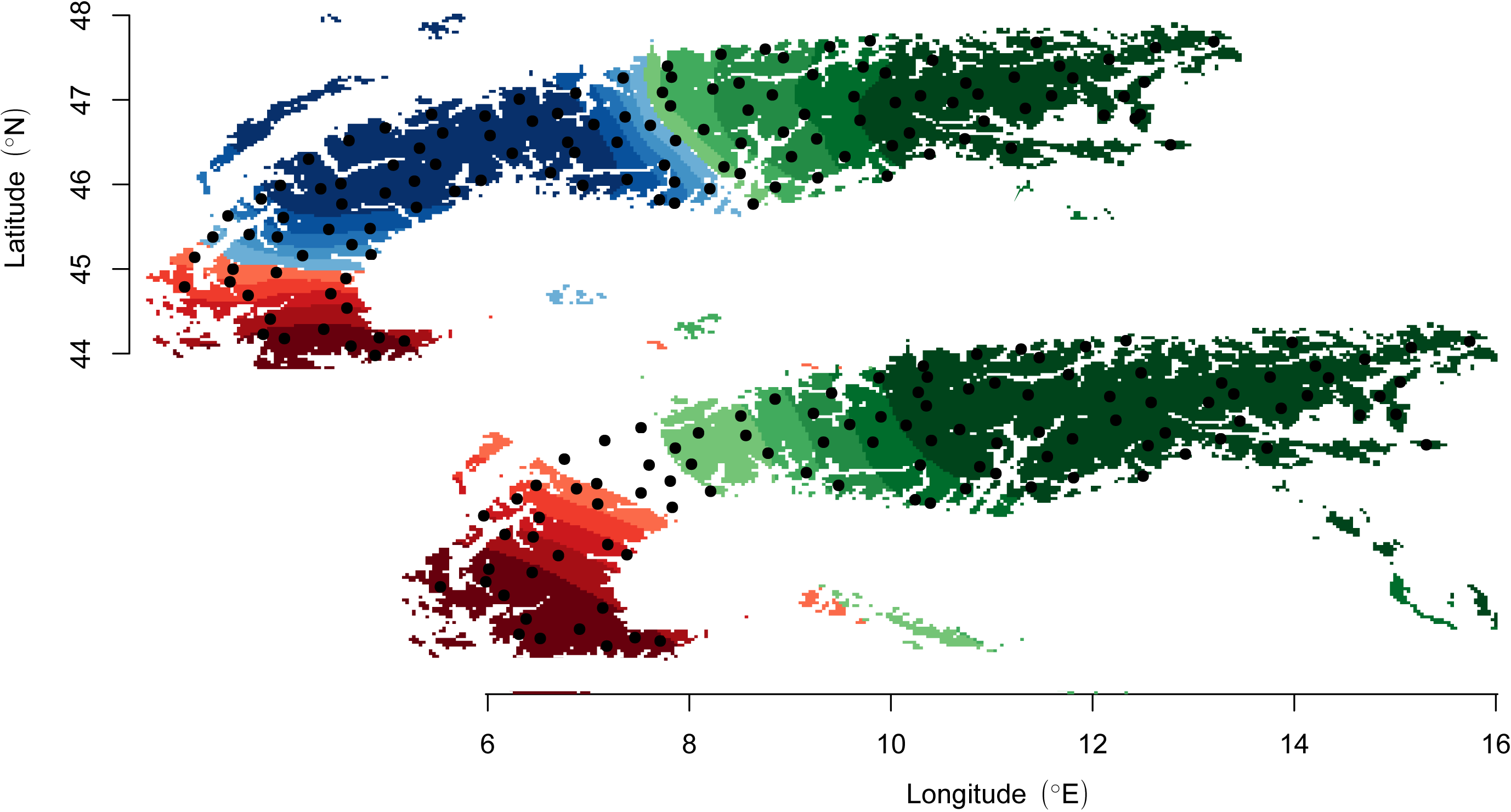
The POPS program performs inference of ancestry distribution models. It uses a TESS-like interface to compute individual cluster membership and admixture proportions based on multilocus genotype data and their correlation with environmental and geographical variables
TESS 2.3: Bayesian Clustering using tessellations and Markov models for spatial population genetics
The POPS program performs inference of ancestry distribution models. It uses a TESS-like interface to compute individual cluster membership and admixture proportions based on multilocus genotype data and their correlation with environmental and geographical variables

没有评论:
发表评论