I collected these guides around the blog posts I found. Please go to the originals for details. And reference should go to the original authors.
1.
Japan Quake Map, with codes and video
library (ggplot2)
library (mapproj)
library (maps)
library (maptools)
# setting parameter about a map.
long <- c (120, 150)
lat <- c (25, 50)
# Reading data of earthquakes and map.
url <- "http://knowledgediscovery.jp/data/eq39.csv"
eq <- read.csv(url, as.is=T)
map <- data.frame(map(xlim = long, ylim = lat))
# creating image with ggplot.
p <- ggplot(eq, aes(long, lat))
p + geom_path(aes (x, y), map) +
geom_point(aes(size = Magnitude, colour = Magnitude), alpha = 1/2) + xlim(long) + ylim(lat)
Created by Pretty R at inside-R.org
2.
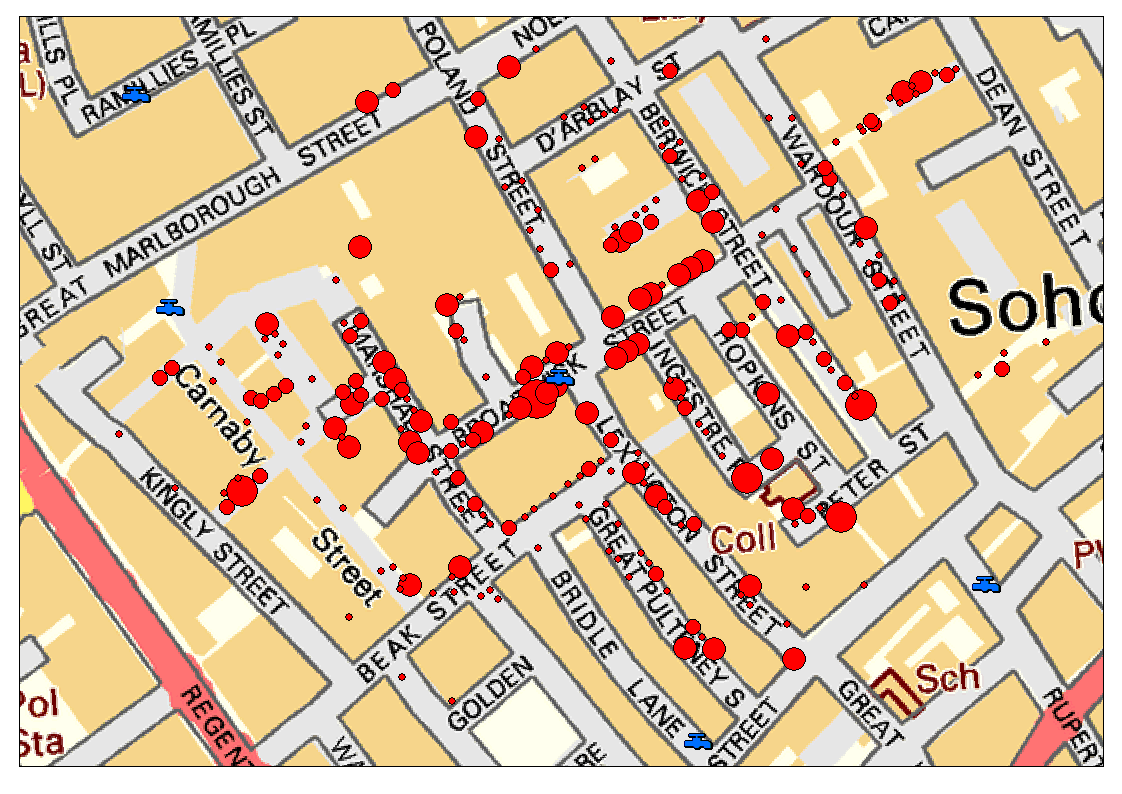
John Snow’s famous cholera analysis data in modern GIS formats, work with shape point format.
Created by Pretty R at inside-R.org
3.
How to draw good looking maps in R,
library(ggplot2)
library(maps)
#load us map data
all_states <- map_data("state")
#plot all states with ggplot
p <- ggplot()
p <- p + geom_polygon( data=all_states, aes(x=long, y=lat, group = group),colour="white", fill="grey10" )
p
Created by Pretty R at inside-R.org
4.
Visualizing GIS data with Rand Open Street Map,
OpenStreetMap, and R package -
RgoogleMaps

# (1) download the map
lat_c<-51.47393
lon_c<-7.22667
bb<-qbbox(lat = c(lat_c[1]+0.01, lat_c[1]-0.01), lon = c(lon_c[1]+0.03, lon_c[1]-0.03))
# download the corresponding Openstreetmap tile with the following line
OSM.map<-GetMap.OSM(lonR=bb$lonR, latR=bb$latR, scale = 20000, destfile=”bochum.png”)
# (2) Add some points to the graphic
lat <- c(51.47393, 51.479021)
lon <- c(7.22667, 7.222526)
val <- c(10, 100)
# As the R-package was mainly build for google-maps, the coordinates need to be adjusted by hand.
# I made the following functions, that take the min and max value from the downloaded map.
lat_adj<-function(lat, map){(map$BBOX$ll[1]-lat)/(map$BBOX$ll[1]-map$BBOX$ur[1])}
lon_adj<-function(lon, map){(map$BBOX$ll[2]-lon)/(map$BBOX$ll[2]-map$BBOX$ur[2])
# (3) add some points to the map. If you want them to mean anything it may be handy to specify
# an alpha-level and change some aspects of the points, e.g. size, color,
# alpha corresponding to some variable of interest.
PlotOnStaticMap(OSM.map, lat = lat_adj(lat, OSM.map), lon = lon_adj(lon, OSM.map), col=rgb(200,val,0,85,maxColorValue=255),pch=16,cex=4)
Created by Pretty R at inside-R.org
5.
Nice Species Distribution Maps with GBIF-Data in R
# go to gbif and download a csv file with the species you
# want (example data)
# ..in R first set working directory to your download directory..
data <- read.csv("occurrence-search-1316347695527.CSV",
header = T, sep = ",")
str(data)
require(maps)
pdf("myr_germ.pdf")
map("world", resolution = 75)
points(data$Longitude, data$Latitude, col = 2, cex = 0.05)
text(-140, -50, "MYRICARIA\nGERMANICA")
dev.off()
Created by Pretty R at inside-R.org
6.
simple example for using shapefiles with R, download
example data.
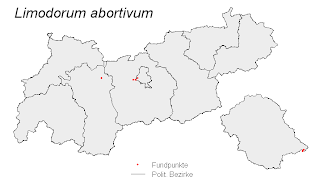
# A simple example for drawing an occurrence-map (polygons with species' points)
# with the R-packages maptools and sp using shapefiles.
library(maptools)
library(sp)
# Note that for each shapefile, you only need to read the .shp component
# the others will be read in at the same time automatically.
# TIRIS.BEZIDX_PL.shp contains the political districts of North-Tyrol
# Limodorum.shp contains points with occurences of the plant species
# Limodorum abortivum.
# Note that my layers use the same geographic coordinate systems (gcs),
# using other data you would need to check if the gcs and
# projection of all layers are the same!
# set dir to where you downloaded the data:
setwd("E:/R/Data/Maps/Example.1_Data")
Limodorum.shp <- readShapePoints(file.choose())
TIRIS.BEZIDX_PL.shp <- readShapePoly(file.choose())
# examine points:
summary(Limodorum.shp)
attributes(Limodorum.shp@data)
# examine polygons:
summary(TIRIS.BEZIDX_PL.shp)
attributes(TIRIS.BEZIDX_PL.shp@data)
# limits:
# to customize ylim in plot-call
# seems not to work here...
xlim <- TIRIS.BEZIDX_PL.shp@bbox[1, ]
ylim <- TIRIS.BEZIDX_PL.shp@bbox[2, ]
par(mai = rep(.1, 4))
plot(TIRIS.BEZIDX_PL.shp, col = "grey93", axes = F,
xlim = xlim, ylim = ylim, bty = "n")
points(Limodorum.shp, pch = 16,
col = 2, cex = .5)
mtext("Limodorum abortivum", 3, line = -7,
at = -17000, adj = 0, cex = 2, font = 3)
legend("bottomleft", inset = c(0.4, 0.2),
legend = c("Fundpunkte", "Polit. Bezirke"),
bty = "n", pch = c(16,-1), # bty = "n": no box
col = c(2, 1), pt.cex = c(.5, 1),
lty = c(-1, 1))
Created by Pretty R at inside-R.org
7.
Weecology can has new mammal dataset
use the rOpenSci package treebase to search the online phylogeny repository TreeBASE. Limiting to returning a max of 1 tree (to save time), wecan see that X species are in at least 1 tree on the TreeBASE database.
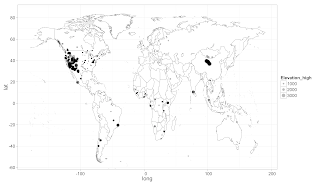
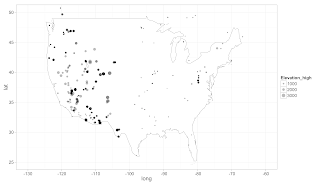
# setwd("/Mac/R_stuff/Blog_etc/Mammal_Dataset")
# URLs for datasets
comm <- "http://esapubs.org/archive/ecol/E092/201/data/MCDB_communities.csv"
refs <- "http://esapubs.org/archive/ecol/E092/201/data/MCDB_references.csv"
sites <- "http://esapubs.org/archive/ecol/E092/201/data/MCDB_sites.csv"
spp <- "http://esapubs.org/archive/ecol/E092/201/data/MCDB_species.csv"
trap <- "http://esapubs.org/archive/ecol/E092/201/data/MCDB_trapping.csv"
# read them
require(plyr)
datasets <- llply(list(comm, refs, sites, spp, trap), read.csv, .progress='text')
str(datasets[[1]]); head(datasets[[1]]) # cool, worked
llply(datasets, head) # see head of all data.frame's
# Map the communities
require(ggplot2)
require(maps)
sitesdata <- datasets[[3]]
sitesdata <- sitesdata[!sitesdata$Latitude == 'NULL',]
sitesdata$Latitude <- as.numeric(as.character(sitesdata$Latitude))
sitesdata$Longitude <- as.numeric(as.character(sitesdata$Longitude))
sitesdata$Elevation_high <- as.numeric(as.character(sitesdata$Elevation_high))
sitesdata <- sitesdata[sitesdata$Longitude > -140,]
world_map <- map_data("world")
us <- world_map[world_map$region=='USA',]
# World map
ggplot(world_map, aes(long, lat)) +
theme_bw(base_size=16) +
geom_polygon(aes(group = group), colour="grey60", fill = 'white', size = .3) +
ylim(-55, 85) +
geom_jitter(data = sitesdata, aes(Longitude, Latitude, size=Elevation_high), alpha=0.3)
ggsave("worldmap.png")
# US only
ggplot(us, aes(long, lat)) +
theme_bw(base_size=16) +
geom_polygon(aes(group = group), colour="grey60", fill = 'white', size = .3) +
ylim(25, 50) +
xlim(-130, -60) +
geom_jitter(data = sitesdata, aes(Longitude, Latitude, size=Elevation_high), alpha=0.3)
ggsave("usmap.png")
# What phylogenies can we get for these species?
install.packages("treebase")
require(treebase); require(RCurl)
datasets[[4]]$gensp <- as.factor(paste(datasets[[4]]$Genus, datasets[[4]]$Species))
trees <- llply(datasets[[4]]$gensp, function(x) search_treebase(paste("\"", x, '\"', sep=''),
by="taxon", max_trees=1, exact_match=TRUE, curl=getCurlHandle()))
spwithtrees <- data.frame(datasets[[4]]$gensp,
laply(trees, function(x) if(length(unlist(x)) > 0){1} else{0}))
names(spwithtrees) <- c('species', 'trees')
length(spwithtrees[spwithtrees$trees == 1, 2])
Created by Pretty R at inside-R.org
8.
googleVis 0.2.13: new stepped area chart and improved geo charts
you should go to the post to see the nice plots.
9.
Spatial Data with R, two videos on how to do
10.
Create maps with maptools R package
library(maptools)
france<-readShapeSpatial("departements.shp",
proj4string=CRS("+proj=longlat"))
routesidf<-readShapeLines(
"ile-de-france.shp/roads.shp",
proj4string=CRS("+proj=longlat")
)
trainsidf<-readShapeLines(
"ile-de-france.shp/railways.shp",
proj4string=CRS("+proj=longlat")
)
plot(france,xlim=c(2.2,2.4),ylim=c(48.75,48.95),lwd=2)
plot(routesidf[routesidf$type=="secondary",],add=TRUE,lwd=2,col="lightgray")
plot(routesidf[routesidf$type=="primary",],add=TRUE,lwd=2,col="lightgray")
plot(trainsidf[trainsidf$type=="rail",],add=TRUE,lwd=1,col="burlywood3")
points(eglises$lon,eglises$lat,pch=20,col="red")
Created by Pretty R at inside-R.org
11.
Producing Google Map Embeds with R Package googleVis
the output plot,
http://localhost:21131/custom/googleVis/MapID3bdbc9a1.html
# (1) for producing html code for a Google Map with R-package googleVis do something like:
library(googleVis)
df <- data.frame(Address = c("Innsbruck", "Wattens"),
Tip = c("My Location 1", "My Location 2"))
mymap <- gvisMap(df, "Address", "Tip", options = list(showTip = TRUE, mapType = "normal",
enableScrollWheel = TRUE))
plot(mymap) # preview
# (2) then just copy-paste the html to your blog or website after customizing for your purpose..
Created by Pretty R at inside-R.org
12.
Retrieve GBIF Species Occurrence Data with Function from dismo Package
The dismo package is awesome: with some short lines of code you can read & map species distribution data from GBIF (theglobal biodiversity information facility)
<div style="overflow:auto;">
library(dismo)
# get GBIF data with function:
myrger <- gbif("Myricaria", "germanica", geo = T)
# check:
str(myrger)
# plot occurrences:
library(maptools)
data(wrld_simpl)
plot(wrld_simpl, col = "light yellow", axes = T)
points(myrger$lon, myrger$lat, col = "red", cex = 0.5)
text(-140, -50, "MYRICARIA\nGERMANICA")
Created by Pretty R at inside-R.org
13.
The Global Earthquake Desktop
The U.S. Geological Survey (USGS) provides feeds and data catalogs of the latestearthquakes, which can be found here.
The maps package in R is great for producing simple maps of data and coding up ourearthquake map is pretty straightforward.
# load the maps library
library(maps)
# get the earthquake data from the USGS
eq <- read.csv("http://earthquake.usgs.gov/earthquakes/catalogs/eqs7day-M2.5.txt", sep = ",", header = TRUE)
# size the earthquake symbol areas according to magnitude
radius <- 10^sqrt(eq$Mag)
# create a time stamp for the image
date <- date()
# set the path and file name of the image file
png("Path/To/Your/Image/worldeq.png", width = 1920, height = 1200)
# plot the world map, earthquakes, and time stamp
map(database = "world",col = "#999999", bg = "#000000")
symbols(eq$Lon, eq$Lat, bg = "#99ccff", fg = "#3467cd", lwd = 1, circles = radius, inches = 0.175, add = TRUE)
text(140, -80, labels = date, col = "#999999")
dev.off()
Created by Pretty R at inside-R.org
# run R in terminal, and automatically update the plot every day.
R CMD Batch /Path/To/Your/R/Script/worldeq.R
defaults write com.apple.desktop Background '{default = {ImageFilePath = "/Path/To/Your/Image/worldeq.png"; };}'
killall Dock
14.
R Spatial Tips
Relevant Book Reviews:
15.
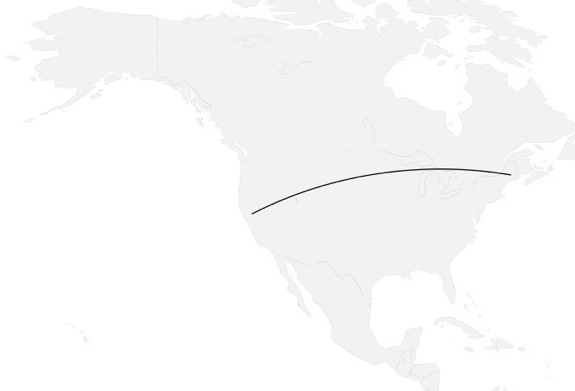
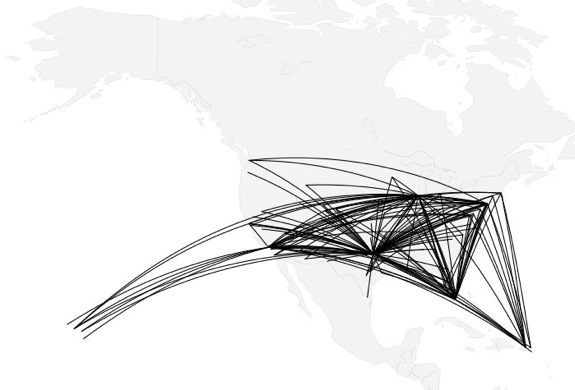
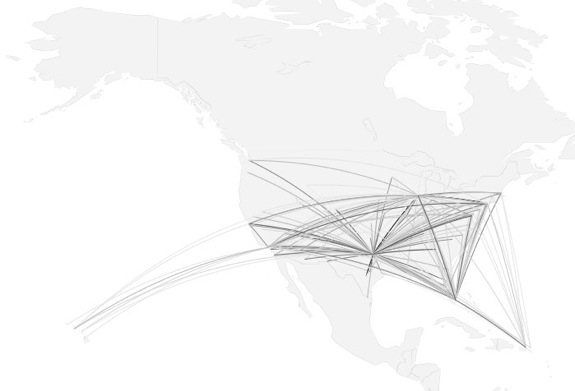
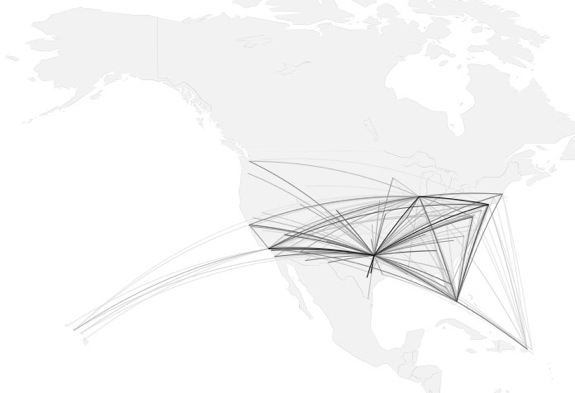
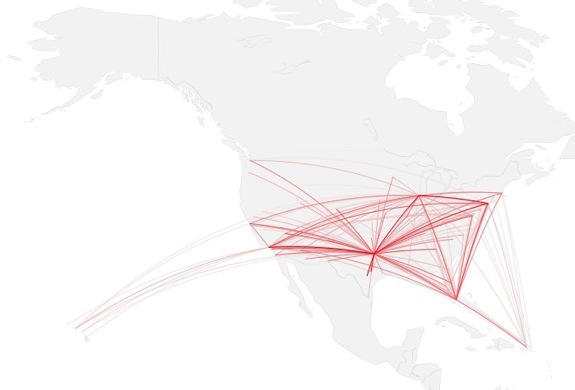
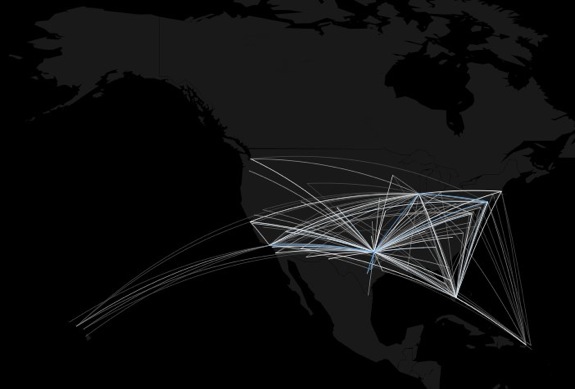
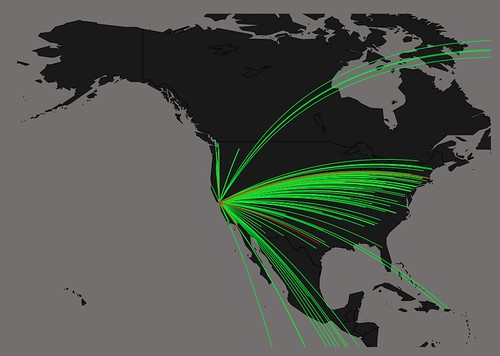
how to map connections with great circles
16.
migration
https://github.com/aihardin/Academic_Migrations/blob/master/map_students.R
#http://flowingdata.com/2011/05/11/how-to-map-connections-with-great-circles
students <- read.csv('/Users/aaron/projects/graduate_student_migrations/students_2005-2011.txt', header=TRUE, as.is=TRUE)
universities <- read.csv('/Users/aaron/projects/graduate_student_migrations/universities_2005-2011.txt', header=TRUE)
library(maps)
library(geosphere)
checkDateLine <- function(l){
n<-0
k<-length(l)
k<-k-1
for (j in 1:k){
n[j] <- l[j+1] - l[j]
}
n <- abs(n)
m<-max(n, rm.na=TRUE)
ifelse(m > 30, TRUE, FALSE)
}
clean.Inter <- function(p1, p2, n, addStartEnd){
inter <- gcIntermediate(p1, p2, n=n, addStartEnd=addStartEnd)
if (checkDateLine(inter[,1])){
m1 <- midPoint(p1, p2)
m1[,1] <- (m1[,1]+180)%%360 - 180
a1 <- antipode(m1)
l1 <- gcIntermediate(p1, a1, n=n, addStartEnd=addStartEnd)
l2 <- gcIntermediate(a1, p2, n=n, addStartEnd=addStartEnd)
l3 <- rbind(l1, l2)
l3
}
else{
inter
}
}
#map("world", col="#191919", fill=TRUE, bg="#736F6E", lwd=0.05)
add_lines <- function(){
pal <- colorRampPalette(c("#00FF00", "#FF0000"))
colors <- pal(100)
fsub <- students[order(students$cnt),]
maxcnt <- max(fsub$cnt)
for (j in 1:length(fsub$undergrad)) {
u1 <- universities[universities$uni_id == fsub[j,]$undergrad,]
u2 <- universities[universities$uni_id == fsub[j,]$grad,]
p1 <- c(u1[1,]$long, u1[1,]$lat)
p2 <- c(u2[1,]$long, u2[1,]$lat)
inter <- clean.Inter(p1,p2,n=100, addStartEnd=TRUE)
colindex <- round( (fsub[j,]$cnt / maxcnt) *length(colors))
lines(inter, col=colors[colindex], lwd=0.6)
}
}
map_usa <- function(){
xlim <- c(-171.738281, -56.601563)
ylim <- c(12.039321, 71.856229)
map("world", col="#191919", fill=TRUE, bg="#736F6E", lwd=0.05, xlim=xlim, ylim=ylim)
add_lines()
}
map_world <- function(){
map("world", col="#191919", fill=TRUE, bg="#736F6E", lwd=0.05)
add_lines()
}
map_usa()
Created by Pretty R at inside-R.org
17.
Amateur Mapmaking: Getting Started With Shapefiles
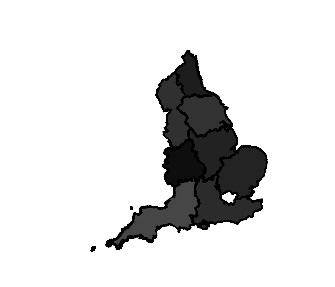
09 | gor=readShapeSpatial('~/Downloads/UK_GORs/Regions.shp') |
13 | insolvency<- read.csv("~/Downloads/csq-q3-2011-insolvency-tables.csv") |
15 | insolvencygor.2011Q3=subset(insolvency,Time.Period=='2011 Q3' & Geography.Type=='Government office region') |
20 | insolvencygor.2011Q3=drop.levels(insolvencygor.2011Q3) |
22 | names(insolvencygor.2011Q3) |
27 | levels(insolvencygor.2011Q3$Geography) |
40 | insolvencygor.2011Q3$Creditors.Petition=as.numeric(levels(insolvencygor.2011Q3$Creditors.Petition))[insolvencygor.2011Q3$Creditors.Petition] |
41 | insolvencygor.2011Q3$Company.Winding.up.Petition=as.numeric(levels(insolvencygor.2011Q3$Company.Winding.up.Petition))[insolvencygor.2011Q3$Company.Winding.up.Petition] |
42 | insolvencygor.2011Q3$Debtors.Petition=as.numeric(levels(insolvencygor.2011Q3$Debtors.Petition))[insolvencygor.2011Q3$Debtors.Petition] |
45 | i2=insolvencygor.2011Q3 |
46 | i2c=c('East of England','East Midlands','Greater London Authority','North East','North West','South East','South West','Wales','West Midlands','Yorkshire and The Humber') |
47 | i2$Geography=factor(i2$Geography,labels=i2c) |
50 | gor@data=merge(gor@data,i2,by.x='NAME',by.y='Geography') |
53 | plot(gor,col=gray(gor@data$Creditors.Petition/max(gor@data$Creditors.Petition))) |
load(url("http://gadm.org/data/rda/JPN_adm2.RData"))
# restricted licence (see: http://www.cs.man.ac.uk/~toby/alan/software/#Licensing)
gpclibPermit()
# which variable identifies polygons?
# looks like ID_2
# convert to a structure ggplot2 can handle...takes a while
japan_map <- fortify(gadm, region = "ID_2")
# join up the useful names
japan_map <- rename(japan_map, c("id" = "ID_2"))
japan_map <- join(japan_map, as.data.frame(gadm)[, c("ID_2", "NAME_1", "NAME_2")])
# a subset for speed
# filled polygons
# everything = SLOW!
# everything, filled polygons
# add points with size defined by populations
+ geom_point(aes(x=long, y=lat, size = population), data=Region)
data=japan_map) + geom_point(aes(size = population), data=Region)
Created by Pretty R at inside-R.org














没有评论:
发表评论